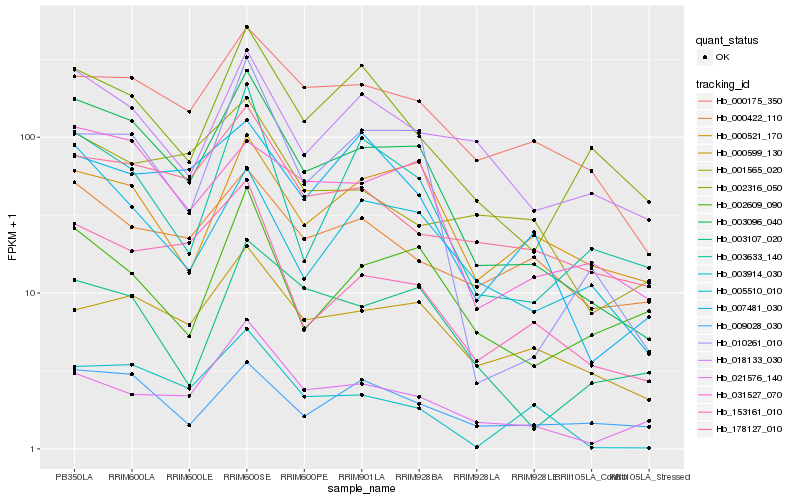
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003096_040 |
0.0 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0005s07290g [Populus trichocarpa] |
| 2 |
Hb_031527_070 |
0.1531007792 |
- |
- |
PREDICTED: isocitrate dehydrogenase [NADP] [Jatropha curcas] |
| 3 |
Hb_010261_010 |
0.1546122792 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 4 |
Hb_153161_010 |
0.1572477406 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 5 |
Hb_000175_350 |
0.1613149503 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 6 |
Hb_178127_010 |
0.1679215097 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 7 |
Hb_005510_010 |
0.1683646659 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X1 [Jatropha curcas] |
| 8 |
Hb_021576_140 |
0.1690501832 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 9 |
Hb_018133_030 |
0.1693526314 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 10 |
Hb_003914_030 |
0.1702072654 |
- |
- |
- |
| 11 |
Hb_003633_140 |
0.1750102584 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000599_130 |
0.1757967169 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 13 |
Hb_002316_050 |
0.177980053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003107_020 |
0.1814131005 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 15 |
Hb_009028_030 |
0.1825492094 |
desease resistance |
Gene Name: NB-ARC |
NBS type disease resistance protein, putative [Theobroma cacao] |
| 16 |
Hb_007481_030 |
0.1870715535 |
- |
- |
PREDICTED: uncharacterized protein LOC105628433 [Jatropha curcas] |
| 17 |
Hb_002609_090 |
0.18836933 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_000521_170 |
0.1910108198 |
- |
- |
PREDICTED: callose synthase 1 [Jatropha curcas] |
| 19 |
Hb_000422_110 |
0.191066775 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 20 |
Hb_001565_020 |
0.1916746692 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |