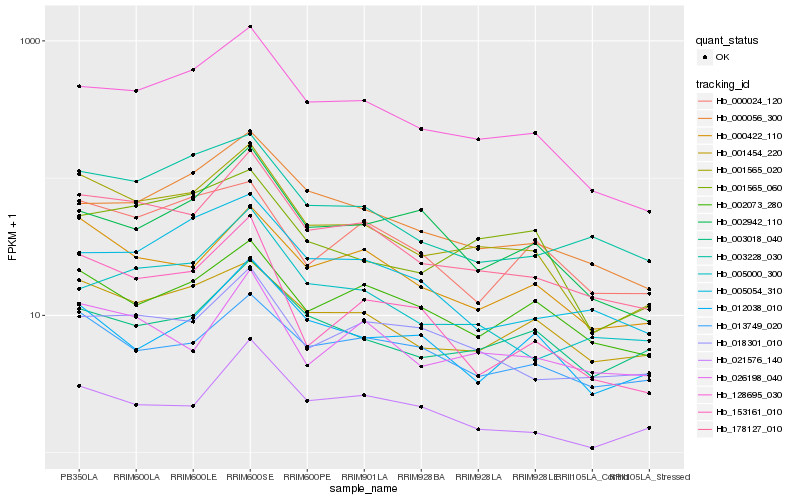
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001565_020 |
0.0 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 2 |
Hb_178127_010 |
0.0801359077 |
- |
- |
Ubiquitin ligase SINAT3, putative [Ricinus communis] |
| 3 |
Hb_128695_030 |
0.089768533 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 4 |
Hb_001454_220 |
0.1215661237 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 5 |
Hb_000422_110 |
0.1220546122 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 6 |
Hb_001565_060 |
0.1283053508 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_153161_010 |
0.128445607 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 8 |
Hb_003228_030 |
0.1336624073 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000056_300 |
0.1375133035 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003018_040 |
0.1436844479 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002073_280 |
0.144878992 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002942_110 |
0.1471432739 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 13 |
Hb_018301_010 |
0.1475804818 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 14 |
Hb_000024_120 |
0.148457428 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 15 |
Hb_013749_020 |
0.1485400853 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_012038_010 |
0.1487146238 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 17 |
Hb_026198_040 |
0.1518050553 |
- |
- |
PREDICTED: uncharacterized protein LOC105628901 [Jatropha curcas] |
| 18 |
Hb_005054_310 |
0.1519195457 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 19 |
Hb_021576_140 |
0.1520017097 |
- |
- |
PREDICTED: protein POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION-like [Jatropha curcas] |
| 20 |
Hb_005000_300 |
0.1532122225 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |