| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
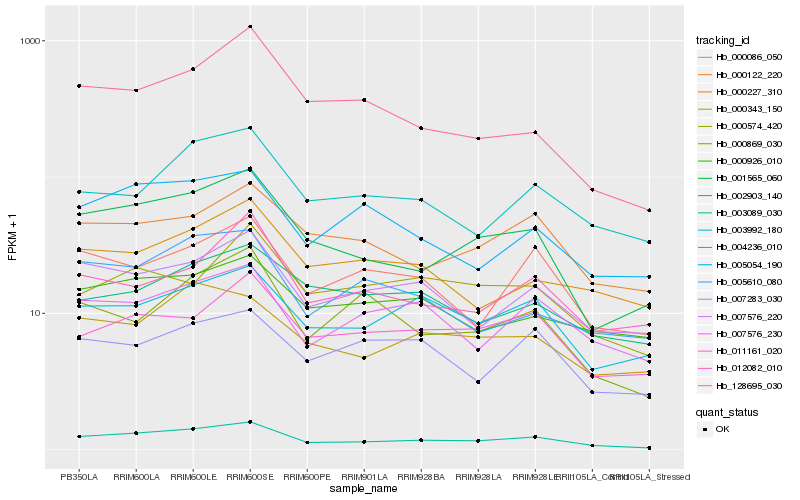
Hb_003089_030 |
0.0 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001565_060 |
0.0891409568 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_007576_220 |
0.1177814843 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 4 |
Hb_004236_010 |
0.11833595 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 5 |
Hb_007576_230 |
0.1226137656 |
- |
- |
hypothetical protein JCGZ_25616 [Jatropha curcas] |
| 6 |
Hb_011161_020 |
0.125292945 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 7 |
Hb_000122_220 |
0.126635333 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 8 |
Hb_000086_050 |
0.1266654059 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 1 [Jatropha curcas] |
| 9 |
Hb_003992_180 |
0.1273426022 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005054_190 |
0.127674712 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000343_150 |
0.1282606107 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 12 |
Hb_000869_030 |
0.1318361895 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |
| 13 |
Hb_128695_030 |
0.1325551348 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 14 |
Hb_005610_080 |
0.1336308744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000926_010 |
0.136731443 |
- |
- |
PREDICTED: uncharacterized protein LOC105632975 [Jatropha curcas] |
| 16 |
Hb_012082_010 |
0.1369491638 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000574_420 |
0.1381123202 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 18 |
Hb_000227_310 |
0.1383285044 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002903_140 |
0.1398547117 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 20 |
Hb_007283_030 |
0.1410723935 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C-like [Malus domestica] |