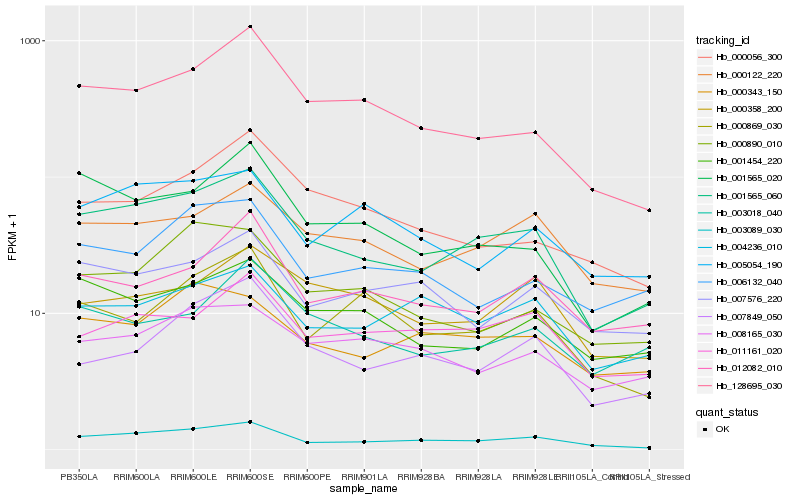
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001565_060 |
0.0 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial [Jatropha curcas] |
| 2 |
Hb_003089_030 |
0.0891409568 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000122_220 |
0.1184339848 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 4 |
Hb_128695_030 |
0.1239235779 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 5 |
Hb_001565_020 |
0.1283053508 |
- |
- |
PREDICTED: probable 3-hydroxyisobutyrate dehydrogenase-like 1, mitochondrial, partial [Jatropha curcas] |
| 6 |
Hb_001454_220 |
0.1289506016 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 7 |
Hb_000343_150 |
0.1412399524 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 8 |
Hb_011161_020 |
0.1414690858 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 9 |
Hb_012082_010 |
0.1415628154 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003018_040 |
0.1420114935 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000890_010 |
0.1447143266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005054_190 |
0.1455770923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007849_050 |
0.1457513314 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 14 |
Hb_007576_220 |
0.1465719224 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 15 |
Hb_004236_010 |
0.1481285063 |
- |
- |
PREDICTED: probable mitochondrial-processing peptidase subunit beta [Jatropha curcas] |
| 16 |
Hb_008165_030 |
0.1490452563 |
- |
- |
PREDICTED: uncharacterized protein LOC105641288 [Jatropha curcas] |
| 17 |
Hb_006132_040 |
0.1499747306 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 18 |
Hb_000358_200 |
0.1513717143 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 19 |
Hb_000056_300 |
0.1518610483 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000869_030 |
0.1519445953 |
transcription factor |
TF Family: Pseudo ARR-B |
PREDICTED: two-component response regulator-like APRR5 isoform X1 [Jatropha curcas] |