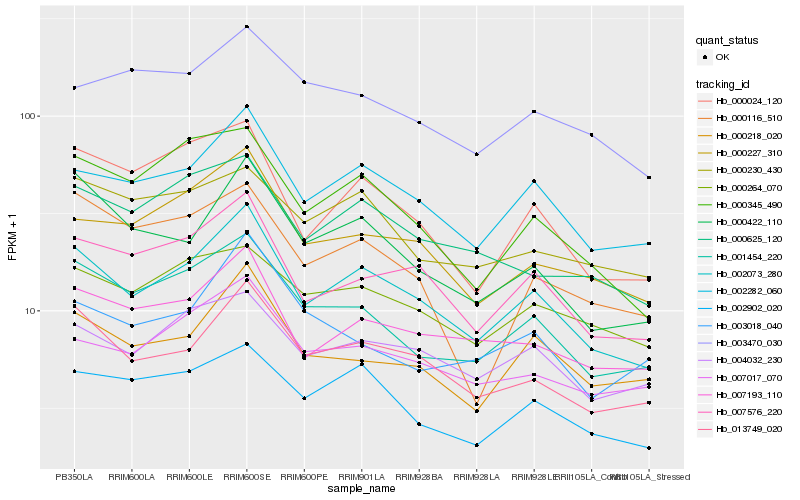
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_220 |
0.0 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 2 |
Hb_000024_120 |
0.0890709142 |
- |
- |
PREDICTED: uncharacterized protein LOC105122546 isoform X1 [Populus euphratica] |
| 3 |
Hb_002073_280 |
0.0892014164 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002902_020 |
0.0908346897 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 5 |
Hb_000345_490 |
0.091105062 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 6 |
Hb_000230_430 |
0.0912432124 |
- |
- |
catalytic, putative [Ricinus communis] |
| 7 |
Hb_007576_220 |
0.0984513356 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 8 |
Hb_000264_070 |
0.1008973674 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 9 |
Hb_007017_070 |
0.1009957351 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000218_020 |
0.1011160665 |
- |
- |
synaptosomal associated protein, putative [Ricinus communis] |
| 11 |
Hb_000422_110 |
0.1013447073 |
- |
- |
PREDICTED: uncharacterized protein At1g08160-like [Jatropha curcas] |
| 12 |
Hb_003018_040 |
0.1019397571 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000227_310 |
0.1023115187 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_002282_060 |
0.1024733726 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_003470_030 |
0.1034985724 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 16 |
Hb_007193_110 |
0.1041363113 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF14 [Jatropha curcas] |
| 17 |
Hb_000625_120 |
0.1042633685 |
- |
- |
PREDICTED: calcium-dependent protein kinase 28 [Jatropha curcas] |
| 18 |
Hb_013749_020 |
0.1048493489 |
- |
- |
PREDICTED: inositol phosphorylceramide glucuronosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_000116_510 |
0.1051288308 |
- |
- |
PREDICTED: RNA-binding protein 24-A-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_004032_230 |
0.1052355041 |
- |
- |
PREDICTED: uncharacterized protein LOC105635143 [Jatropha curcas] |