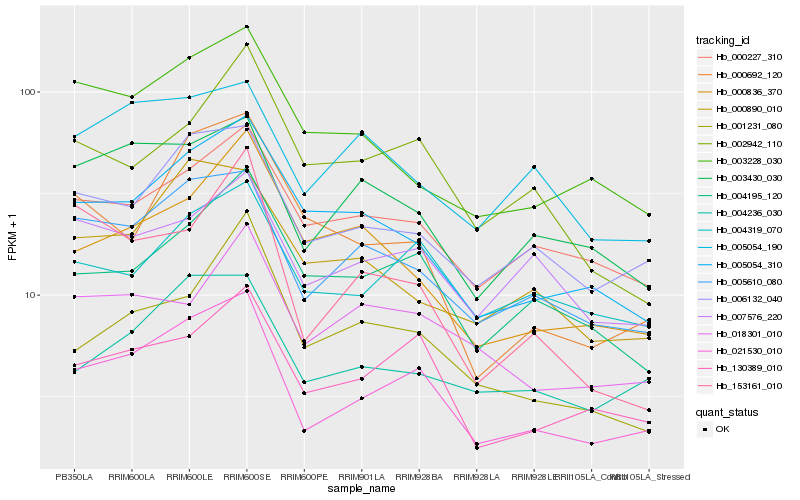
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021530_010 |
0.0 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 2 |
Hb_005610_080 |
0.1052348488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006132_040 |
0.1108201996 |
- |
- |
hypothetical protein JCGZ_14390 [Jatropha curcas] |
| 4 |
Hb_130389_010 |
0.1112901173 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 5 |
Hb_004236_030 |
0.1261168562 |
- |
- |
unknown [Lotus japonicus] |
| 6 |
Hb_005054_310 |
0.1307799441 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 7 |
Hb_003430_030 |
0.1317979774 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp31 [Jatropha curcas] |
| 8 |
Hb_004319_070 |
0.1326186805 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 9 |
Hb_004195_120 |
0.1332754989 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 10 |
Hb_000227_310 |
0.135717044 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_003228_030 |
0.1440629941 |
- |
- |
PREDICTED: transmembrane and coiled-coil domain-containing protein 1 [Jatropha curcas] |
| 12 |
Hb_000890_010 |
0.144125538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007576_220 |
0.1466293474 |
- |
- |
PREDICTED: kanadaptin [Jatropha curcas] |
| 14 |
Hb_001231_080 |
0.1485784204 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 15 |
Hb_018301_010 |
0.1518942757 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 16 |
Hb_000692_120 |
0.1520451353 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 17 |
Hb_000836_370 |
0.1529366629 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_005054_190 |
0.1537481991 |
- |
- |
PREDICTED: uncharacterized protein LOC105646630 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002942_110 |
0.1542338107 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 2-like [Jatropha curcas] |
| 20 |
Hb_153161_010 |
0.1545536829 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |