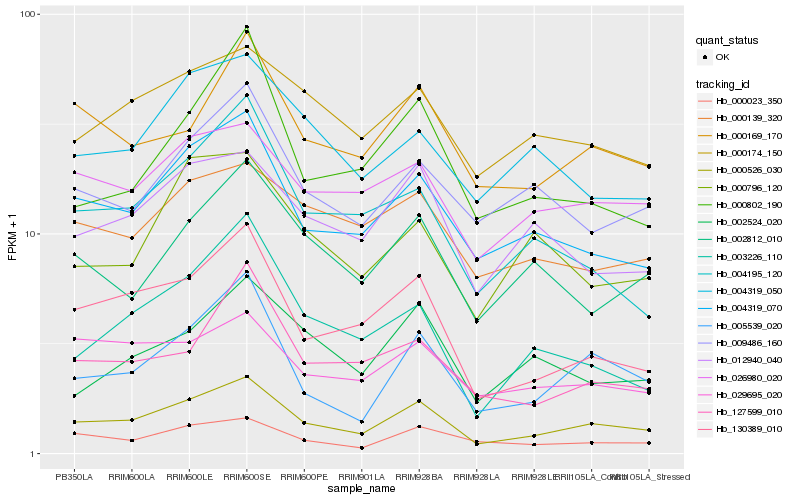
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000526_030 |
0.0 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 2 |
Hb_004319_070 |
0.1009163192 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 3 |
Hb_130389_010 |
0.1013002112 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 4 |
Hb_005539_020 |
0.1039463194 |
- |
- |
- |
| 5 |
Hb_000023_350 |
0.1161093847 |
- |
- |
PREDICTED: probable folate-biopterin transporter 6 isoform X2 [Populus euphratica] |
| 6 |
Hb_000169_170 |
0.1190371742 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 7 |
Hb_012940_040 |
0.1214001367 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |
| 8 |
Hb_026980_020 |
0.1224036692 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 9 |
Hb_000174_150 |
0.1279247543 |
- |
- |
stearoyl-acyl-carrier protein desaturase [Ricinus communis] |
| 10 |
Hb_009486_160 |
0.1282440934 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 11 |
Hb_002812_010 |
0.1287770173 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_127599_010 |
0.1289010747 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 13 |
Hb_004195_120 |
0.130128531 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 14 |
Hb_029695_020 |
0.1313789126 |
- |
- |
PREDICTED: inactive beta-amylase 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000802_190 |
0.1329614057 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 16 |
Hb_003226_110 |
0.1351521593 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 17 |
Hb_000139_320 |
0.1351655465 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 18 |
Hb_004319_050 |
0.1358461804 |
- |
- |
PREDICTED: senescence-associated carboxylesterase 101-like [Populus euphratica] |
| 19 |
Hb_002524_020 |
0.1363412669 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_000796_120 |
0.1369134777 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |