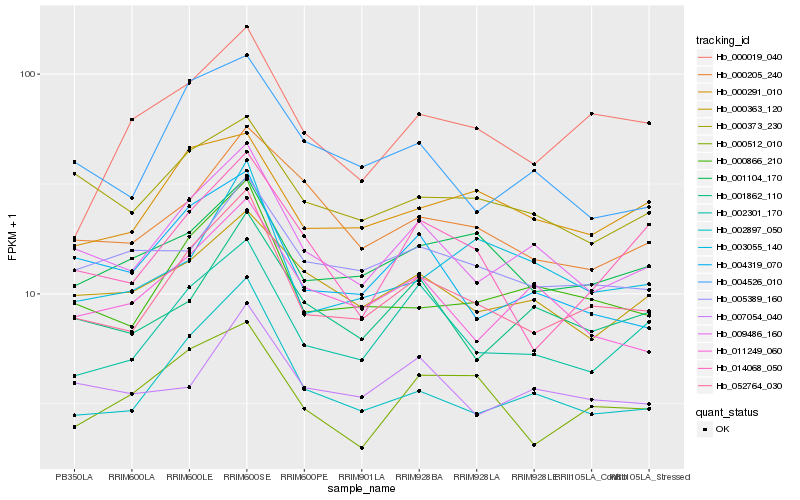
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052764_030 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 2 |
Hb_000866_210 |
0.0781572364 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009486_160 |
0.0856913095 |
- |
- |
PREDICTED: metacaspase-1 [Jatropha curcas] |
| 4 |
Hb_002897_050 |
0.0876137572 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 5 |
Hb_002301_170 |
0.0885910292 |
- |
- |
MAC/Perforin domain-containing protein [Theobroma cacao] |
| 6 |
Hb_001104_170 |
0.0996614798 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 7 |
Hb_000291_010 |
0.10511558 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 8 |
Hb_005389_160 |
0.1092275965 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 9 |
Hb_000512_010 |
0.1093907463 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 10 |
Hb_001862_110 |
0.1119018914 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 11 |
Hb_004319_070 |
0.1119184748 |
- |
- |
hypothetical protein RCOM_0852460 [Ricinus communis] |
| 12 |
Hb_000373_230 |
0.1119712204 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 13 |
Hb_000205_240 |
0.1135450269 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_000019_040 |
0.1148219463 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 15 |
Hb_004526_010 |
0.1162300741 |
- |
- |
signal recognition particle receptor alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_007054_040 |
0.1165451279 |
- |
- |
PREDICTED: protein RST1 [Jatropha curcas] |
| 17 |
Hb_014068_050 |
0.1165988849 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase RLCKVII [Jatropha curcas] |
| 18 |
Hb_003055_140 |
0.116893326 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 19 |
Hb_011249_060 |
0.1170995657 |
- |
- |
hypothetical protein POPTR_0018s12600g [Populus trichocarpa] |
| 20 |
Hb_000363_120 |
0.1173124552 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |