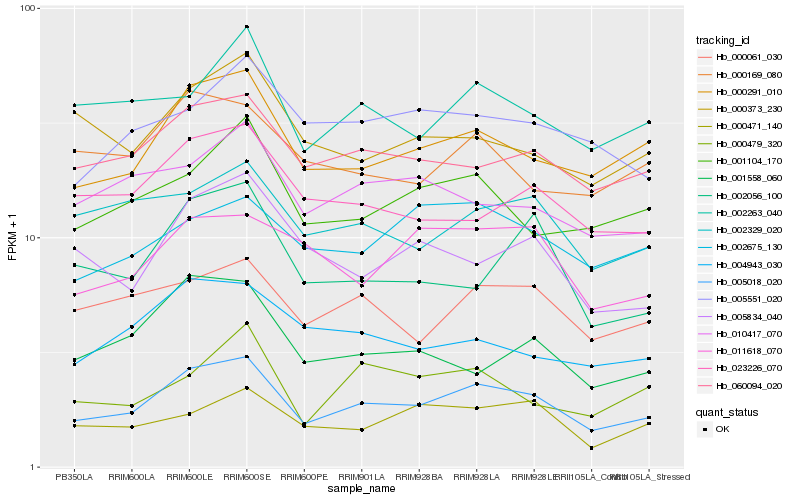
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005018_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000291_010 |
0.08403777 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 3 |
Hb_060094_020 |
0.1017048173 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 4 |
Hb_000061_030 |
0.1096275164 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065 [Jatropha curcas] |
| 5 |
Hb_001104_170 |
0.1096434835 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 6 |
Hb_002263_040 |
0.1122707005 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 7 |
Hb_002329_020 |
0.1144613003 |
- |
- |
PREDICTED: uncharacterized protein LOC105648015 [Jatropha curcas] |
| 8 |
Hb_023226_070 |
0.1171821901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_010417_070 |
0.1189906143 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005834_040 |
0.120146405 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 11 |
Hb_004943_030 |
0.1205659222 |
- |
- |
PREDICTED: cation-chloride cotransporter 1 [Jatropha curcas] |
| 12 |
Hb_011618_070 |
0.1206517741 |
- |
- |
NMDA receptor-regulated protein [Medicago truncatula] |
| 13 |
Hb_000471_140 |
0.1213914653 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g28640 [Prunus mume] |
| 14 |
Hb_005551_020 |
0.1217858931 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 15 |
Hb_002056_100 |
0.1220654721 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 16 |
Hb_002675_130 |
0.122188379 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 16, NatA auxiliary subunit isoform X1 [Jatropha curcas] |
| 17 |
Hb_001558_060 |
0.1223103195 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000373_230 |
0.1227798094 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 19 |
Hb_000169_080 |
0.1234400329 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 3 [Jatropha curcas] |
| 20 |
Hb_000479_320 |
0.1240680714 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06140, mitochondrial-like [Jatropha curcas] |