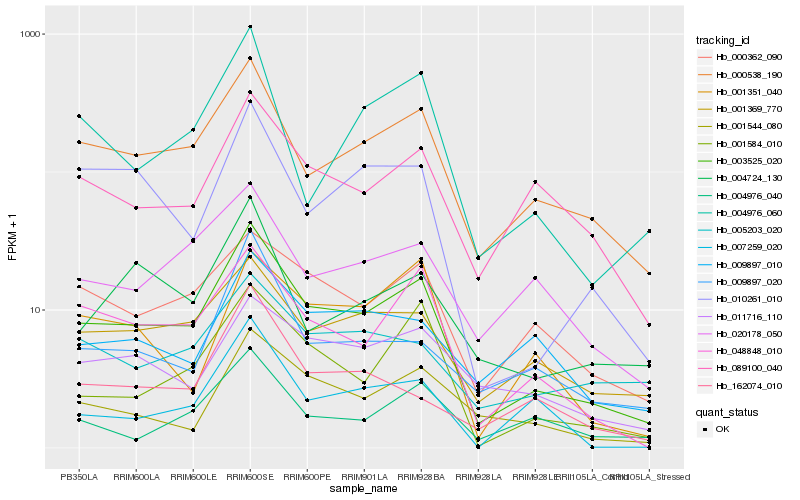
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003525_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000538_190 |
0.1192555509 |
- |
- |
PREDICTED: protein DJ-1 homolog D [Jatropha curcas] |
| 3 |
Hb_048848_010 |
0.1382525527 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 4 |
Hb_001369_770 |
0.1558942102 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 5 |
Hb_001584_010 |
0.1600427322 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 6 |
Hb_010261_010 |
0.1639445948 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX3, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000362_090 |
0.1641091301 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 8 |
Hb_089100_040 |
0.1662108638 |
- |
- |
PREDICTED: uncharacterized protein LOC105638961 [Jatropha curcas] |
| 9 |
Hb_004976_060 |
0.1685741097 |
- |
- |
- |
| 10 |
Hb_001544_080 |
0.170215707 |
- |
- |
PREDICTED: 66 kDa stress protein-like [Jatropha curcas] |
| 11 |
Hb_020178_050 |
0.1765923023 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 12 |
Hb_011716_110 |
0.1771130491 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 13 |
Hb_004724_130 |
0.1813662728 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 14 |
Hb_007259_020 |
0.1817234433 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 [Jatropha curcas] |
| 15 |
Hb_009897_020 |
0.1837514184 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 16 |
Hb_009897_010 |
0.18558381 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_162074_010 |
0.1860779186 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 18 |
Hb_005203_020 |
0.1882775952 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 19 |
Hb_001351_040 |
0.1899651829 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_004976_040 |
0.1911360064 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |