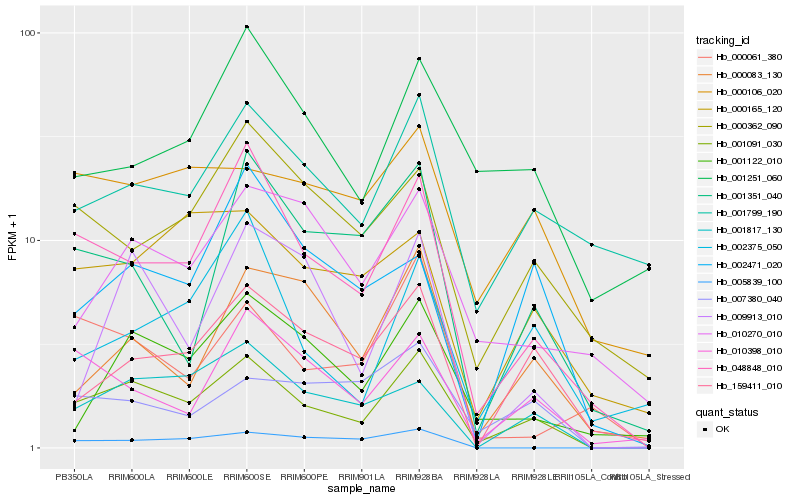
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001091_030 |
0.0 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |
| 2 |
Hb_048848_010 |
0.1493013752 |
- |
- |
hypothetical protein POPTR_0005s07810g [Populus trichocarpa] |
| 3 |
Hb_001817_130 |
0.1777989122 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 isoform X3 [Cucumis melo] |
| 4 |
Hb_010398_010 |
0.2009444694 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: mediator of RNA polymerase II transcription subunit 15a-like [Malus domestica] |
| 5 |
Hb_159411_010 |
0.2012309852 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 6 |
Hb_000362_090 |
0.2120851614 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007380_040 |
0.2124187876 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 8 |
Hb_000165_120 |
0.2124397141 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 9 |
Hb_001122_010 |
0.2124938101 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_000106_020 |
0.2143205322 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 11 |
Hb_001351_040 |
0.2169278653 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_001799_190 |
0.219951959 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 13 |
Hb_002375_050 |
0.220547336 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 14 |
Hb_000083_130 |
0.2213959746 |
- |
- |
hypothetical protein RCOM_0884570 [Ricinus communis] |
| 15 |
Hb_009913_010 |
0.2231131712 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000061_380 |
0.2238896188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010270_010 |
0.2247002541 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002471_020 |
0.2262148835 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 6-like [Jatropha curcas] |
| 19 |
Hb_001251_060 |
0.2294780029 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 20 |
Hb_005839_100 |
0.2296146902 |
- |
- |
PREDICTED: APO protein 1, chloroplastic isoform X3 [Jatropha curcas] |