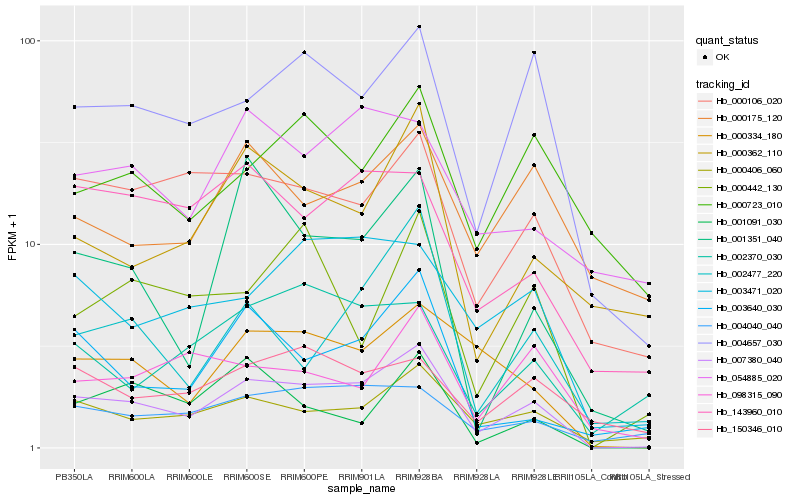
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007380_040 |
0.0 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 2 |
Hb_004657_030 |
0.1570426116 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX14 [Jatropha curcas] |
| 3 |
Hb_000406_060 |
0.1614689908 |
- |
- |
hypothetical protein CICLE_v10000115mg [Citrus clementina] |
| 4 |
Hb_000106_020 |
0.1777653887 |
- |
- |
PREDICTED: protein TOPLESS isoform X1 [Jatropha curcas] |
| 5 |
Hb_003471_020 |
0.1811081382 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Jatropha curcas] |
| 6 |
Hb_000334_180 |
0.1814882308 |
- |
- |
hypothetical protein JCGZ_16779 [Jatropha curcas] |
| 7 |
Hb_004040_040 |
0.1885423959 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 8 |
Hb_143960_010 |
0.1944833471 |
- |
- |
- |
| 9 |
Hb_003640_030 |
0.1968738457 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002477_220 |
0.1974586526 |
- |
- |
hypothetical protein POPTR_0017s06650g [Populus trichocarpa] |
| 11 |
Hb_000362_110 |
0.2000754569 |
- |
- |
PREDICTED: uncharacterized protein LOC104612097 isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_150346_010 |
0.2030668446 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 13 |
Hb_000442_130 |
0.2034095079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001351_040 |
0.2053942833 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_098315_090 |
0.2057956105 |
- |
- |
Aldehyde dehydrogenase family 6 member B2 [Morus notabilis] |
| 16 |
Hb_002370_030 |
0.210047075 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 17 |
Hb_000723_010 |
0.210661663 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_000175_120 |
0.2108610503 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 19 |
Hb_054885_020 |
0.2118178876 |
- |
- |
- |
| 20 |
Hb_001091_030 |
0.2124187876 |
- |
- |
hypothetical protein AALP_AA1G173500 [Arabis alpina] |