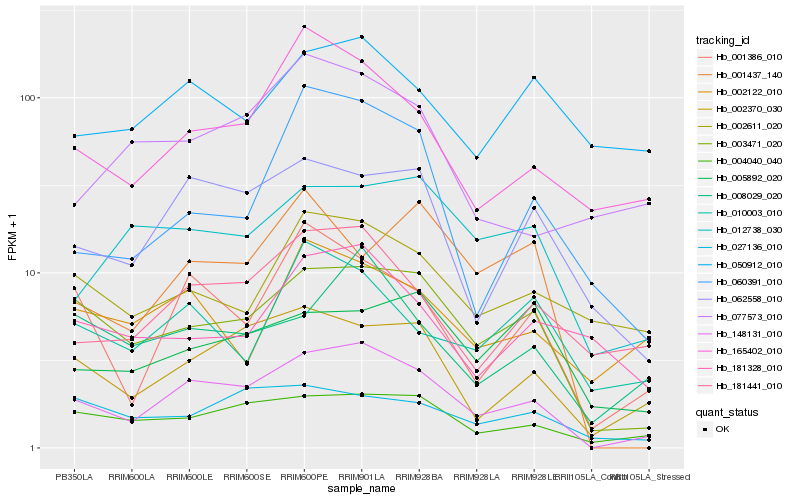
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148131_010 |
0.0 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12-like, partial [Jatropha curcas] |
| 2 |
Hb_003471_020 |
0.1352715086 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8A [Jatropha curcas] |
| 3 |
Hb_002370_030 |
0.1542433439 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 4 |
Hb_004040_040 |
0.1617705344 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 5 |
Hb_001386_010 |
0.1657981456 |
- |
- |
PREDICTED: uncharacterized protein LOC105632444 [Jatropha curcas] |
| 6 |
Hb_062558_010 |
0.1675105223 |
- |
- |
PREDICTED: L-2-hydroxyglutarate dehydrogenase, mitochondrial isoform X1 [Populus euphratica] |
| 7 |
Hb_181441_010 |
0.1754186901 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 8 |
Hb_008029_020 |
0.1819449064 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 9 |
Hb_165402_010 |
0.1852172757 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 10 |
Hb_012738_030 |
0.1960989866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002611_020 |
0.199779136 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 12 |
Hb_027136_010 |
0.2041031364 |
- |
- |
hypothetical protein L484_010426 [Morus notabilis] |
| 13 |
Hb_060391_010 |
0.2090142832 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_002122_010 |
0.2103495571 |
- |
- |
- |
| 15 |
Hb_010003_010 |
0.2121446532 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 16 |
Hb_050912_010 |
0.2122529929 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_005892_020 |
0.2130037427 |
- |
- |
PREDICTED: uncharacterized protein LOC105632170 isoform X1 [Jatropha curcas] |
| 18 |
Hb_077573_010 |
0.2152443036 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_181328_010 |
0.2152616068 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001437_140 |
0.2155588816 |
- |
- |
- |