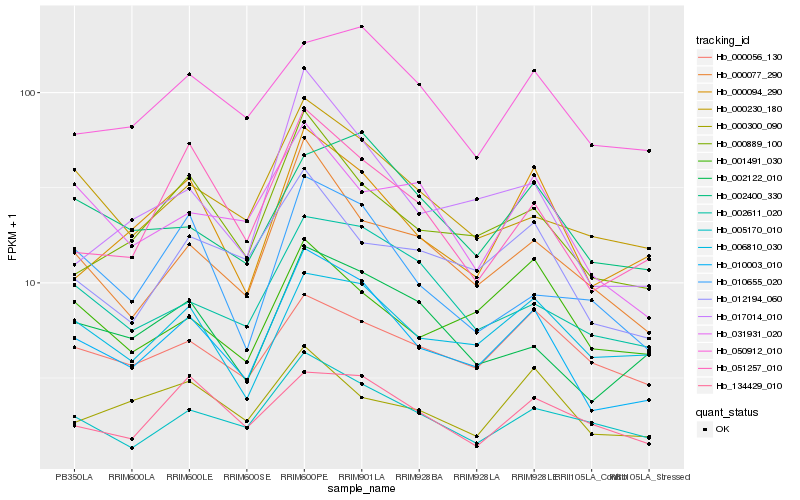
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010003_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000889_100 |
0.1216258212 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 3 |
Hb_000094_290 |
0.1234127208 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000077_290 |
0.1255524555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000230_180 |
0.12678766 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_051257_010 |
0.1299237393 |
- |
- |
- |
| 7 |
Hb_005170_010 |
0.1303305278 |
- |
- |
- |
| 8 |
Hb_002122_010 |
0.1304594545 |
- |
- |
- |
| 9 |
Hb_012194_060 |
0.1345984116 |
- |
- |
PREDICTED: uncharacterized protein LOC105636108 [Jatropha curcas] |
| 10 |
Hb_017014_010 |
0.1351077911 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_006810_030 |
0.1352188683 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_002611_020 |
0.1400204449 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 13 |
Hb_010655_020 |
0.1435871842 |
- |
- |
hypothetical protein CICLE_v10032410mg [Citrus clementina] |
| 14 |
Hb_002400_330 |
0.1456425603 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 15 |
Hb_031931_020 |
0.1468987942 |
- |
- |
Minor histocompatibility antigen H13, putative [Ricinus communis] |
| 16 |
Hb_050912_010 |
0.1473461472 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001491_030 |
0.1482359014 |
- |
- |
PREDICTED: uncharacterized protein LOC105649804 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000300_090 |
0.1511750426 |
- |
- |
catalytic, putative [Ricinus communis] |
| 19 |
Hb_000056_130 |
0.1529966276 |
- |
- |
Sucrose Transporter 2C [Hevea brasiliensis subsp. brasiliensis] |
| 20 |
Hb_134429_010 |
0.1535895063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |