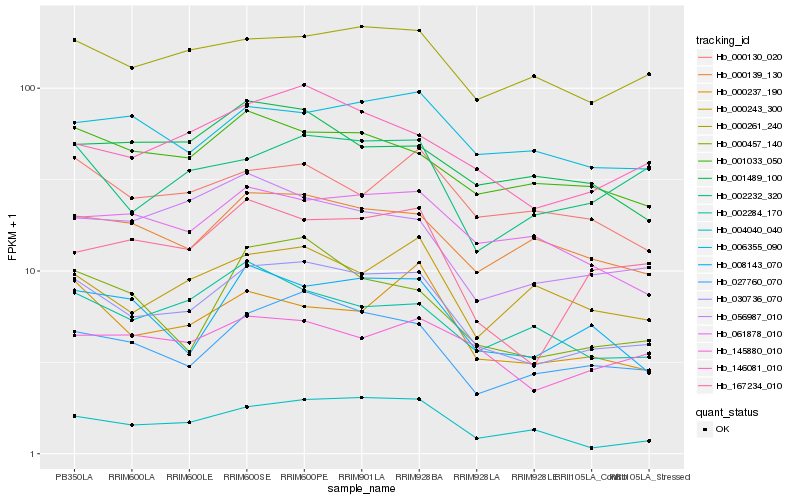
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030736_070 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_027760_070 |
0.0856825048 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 3 |
Hb_146081_010 |
0.0964282589 |
- |
- |
PREDICTED: protein SCAI [Jatropha curcas] |
| 4 |
Hb_008143_070 |
0.1076836835 |
- |
- |
PREDICTED: factor of DNA methylation 4 [Jatropha curcas] |
| 5 |
Hb_001033_050 |
0.1079986832 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 6 |
Hb_000457_140 |
0.1095256149 |
- |
- |
PREDICTED: transmembrane protein 136-like [Jatropha curcas] |
| 7 |
Hb_000237_190 |
0.1104256861 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000139_130 |
0.1113280065 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 9 |
Hb_056987_010 |
0.1130935678 |
- |
- |
DNA-damage-repair/toleration protein DRT111, chloroplastic [Glycine soja] |
| 10 |
Hb_000261_240 |
0.1150643621 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 11 |
Hb_167234_010 |
0.1156431558 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 12 |
Hb_145880_010 |
0.117729143 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 13 |
Hb_002232_320 |
0.1178534638 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_061878_010 |
0.1180734309 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 15 |
Hb_004040_040 |
0.1196344864 |
desease resistance |
Gene Name: ABC_tran |
hypothetical protein POPTR_0006s07370g [Populus trichocarpa] |
| 16 |
Hb_002284_170 |
0.1201734017 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1b [Jatropha curcas] |
| 17 |
Hb_006355_090 |
0.1233276093 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 18 |
Hb_000130_020 |
0.123345213 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 19 |
Hb_001489_100 |
0.1241377587 |
- |
- |
PREDICTED: hypersensitive-induced response protein 2 [Jatropha curcas] |
| 20 |
Hb_000243_300 |
0.1244894083 |
- |
- |
catalytic, putative [Ricinus communis] |