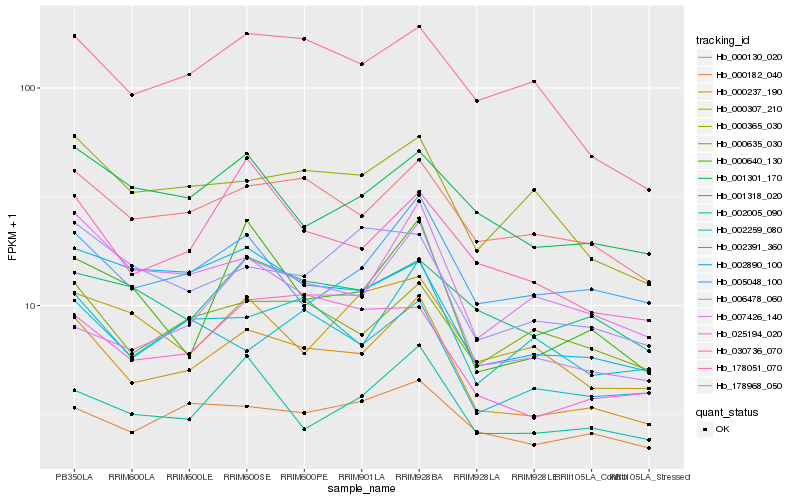
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_190 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000130_020 |
0.0926122865 |
- |
- |
Phosphoenolpyruvate carboxylase, putative [Ricinus communis] |
| 3 |
Hb_025194_020 |
0.1029227393 |
- |
- |
PREDICTED: uncharacterized protein C1F8.04c-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_002259_080 |
0.1035522901 |
- |
- |
protein transport protein sec23, putative [Ricinus communis] |
| 5 |
Hb_000365_030 |
0.1071341243 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 6 |
Hb_178968_050 |
0.1087793613 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 7 |
Hb_030736_070 |
0.1104256861 |
- |
- |
PREDICTED: general transcription factor IIH subunit 3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005048_100 |
0.1104737458 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 9 |
Hb_002005_090 |
0.1106424653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000635_030 |
0.1115760955 |
- |
- |
mannosyl-oligosaccharide glucosidase, putative [Ricinus communis] |
| 11 |
Hb_001301_170 |
0.1136738125 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 12 |
Hb_002391_360 |
0.113781475 |
- |
- |
hypothetical protein CICLE_v10026208mg [Citrus clementina] |
| 13 |
Hb_006478_060 |
0.1147422364 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_178051_070 |
0.1148184201 |
- |
- |
PREDICTED: exocyst complex component EXO70B1-like [Jatropha curcas] |
| 15 |
Hb_002890_100 |
0.1174499063 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein MAD1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000640_130 |
0.1189448644 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001318_020 |
0.1207572377 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 18 |
Hb_000182_040 |
0.1207628333 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 19 |
Hb_007426_140 |
0.1213125177 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 20 |
Hb_000307_210 |
0.1219789265 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |