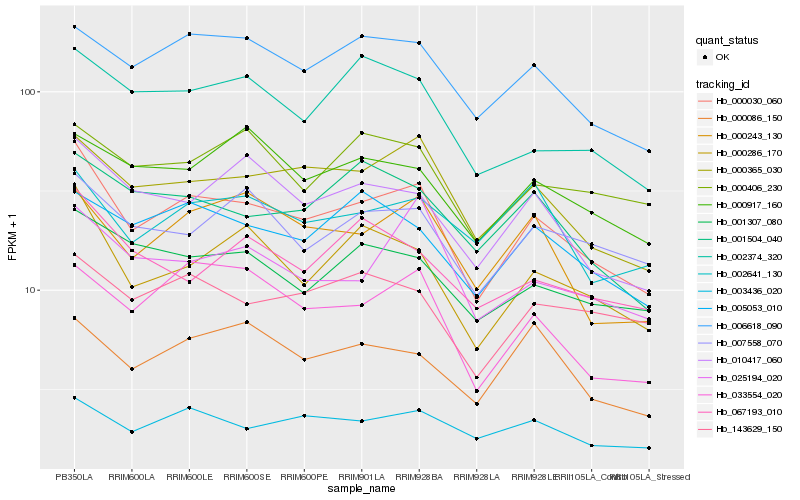
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_060 |
0.0 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 2 |
Hb_000365_030 |
0.0871155145 |
- |
- |
PREDICTED: transmembrane protein 87B [Jatropha curcas] |
| 3 |
Hb_000286_170 |
0.0883811919 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 4 |
Hb_006618_090 |
0.0970099631 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 5 |
Hb_010417_060 |
0.0996762364 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 6 |
Hb_001307_080 |
0.1043783787 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 7 |
Hb_033554_020 |
0.107441029 |
transcription factor |
TF Family: BES1 |
PREDICTED: beta-amylase 8 [Jatropha curcas] |
| 8 |
Hb_007558_070 |
0.1076323634 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005053_010 |
0.1080457941 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 10 |
Hb_000243_130 |
0.1089827555 |
- |
- |
hypothetical protein M569_10795, partial [Genlisea aurea] |
| 11 |
Hb_003436_020 |
0.1089866141 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002374_320 |
0.1105576949 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 13 |
Hb_143629_150 |
0.1114435984 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 14 |
Hb_001504_040 |
0.1117962683 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 15 |
Hb_067193_010 |
0.1152770011 |
- |
- |
- |
| 16 |
Hb_000917_160 |
0.1155849399 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_025194_020 |
0.1156814537 |
- |
- |
PREDICTED: uncharacterized protein C1F8.04c-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000406_230 |
0.1166398876 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 19 |
Hb_002641_130 |
0.1170356216 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 20 |
Hb_000086_150 |
0.1170477683 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X1 [Jatropha curcas] |