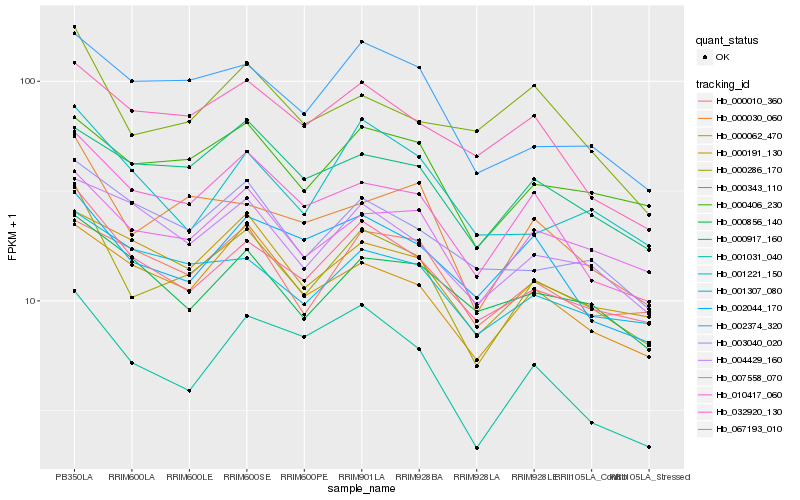
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000286_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 2 |
Hb_067193_010 |
0.0830188138 |
- |
- |
- |
| 3 |
Hb_000030_060 |
0.0883811919 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 4 |
Hb_007558_070 |
0.0950253103 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 5 |
Hb_010417_060 |
0.0983190345 |
- |
- |
PREDICTED: protein SGT1 homolog A-like [Jatropha curcas] |
| 6 |
Hb_000406_230 |
0.1015250706 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 7 |
Hb_000062_470 |
0.1030061887 |
- |
- |
PREDICTED: cyclin-dependent kinase G-2 [Jatropha curcas] |
| 8 |
Hb_002374_320 |
0.1041496172 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 9 |
Hb_000191_130 |
0.1107121645 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 10 |
Hb_001031_040 |
0.1110723658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000917_160 |
0.1125516262 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001307_080 |
0.1126280115 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_002044_170 |
0.1129927896 |
- |
- |
hypothetical protein POPTR_0014s17160g [Populus trichocarpa] |
| 14 |
Hb_000343_110 |
0.1139648158 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 15 |
Hb_003040_020 |
0.1140871676 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 16 |
Hb_004429_160 |
0.1144011866 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001221_150 |
0.1145718225 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 18 |
Hb_000010_360 |
0.1171236616 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 19 |
Hb_000856_140 |
0.1188561159 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 20 |
Hb_032920_130 |
0.1190714972 |
- |
- |
PREDICTED: vacuolar-sorting receptor 3 [Jatropha curcas] |