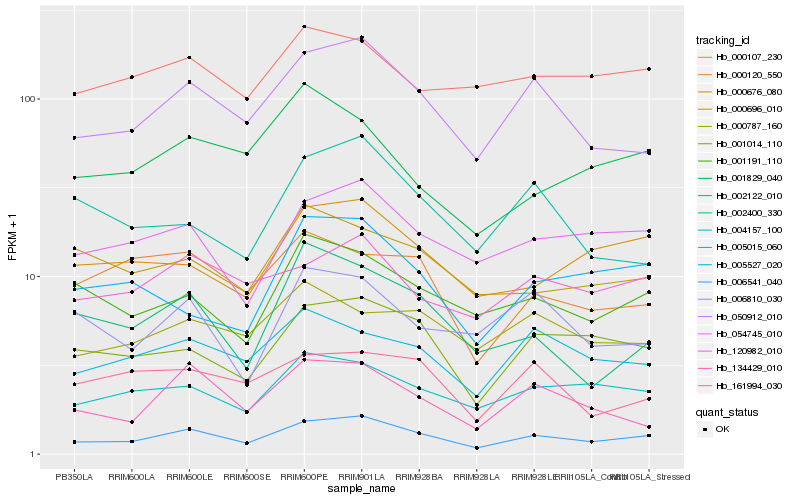
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006541_040 |
0.0 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 2 |
Hb_050912_010 |
0.0952147668 |
- |
- |
PREDICTED: amidase 1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001014_110 |
0.1095481903 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 4 |
Hb_054745_010 |
0.1143865751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004157_100 |
0.1193528366 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 6 |
Hb_000676_080 |
0.127800852 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 7 |
Hb_005015_060 |
0.128288875 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Jatropha curcas] |
| 8 |
Hb_120982_010 |
0.1284934322 |
- |
- |
hypothetical protein CICLE_v10003116mg, partial [Citrus clementina] |
| 9 |
Hb_134429_010 |
0.1287340706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001191_110 |
0.1292229563 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 11 |
Hb_005527_020 |
0.1399589319 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 12 |
Hb_000696_010 |
0.1440138873 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 13 |
Hb_000107_230 |
0.1448779113 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_000787_160 |
0.1460117924 |
- |
- |
PREDICTED: GDP-mannose transporter GONST1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002122_010 |
0.1460203699 |
- |
- |
- |
| 16 |
Hb_000120_550 |
0.1465575491 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 17 |
Hb_161994_030 |
0.14703604 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_002400_330 |
0.147103366 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 19 |
Hb_001829_040 |
0.1472322784 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 20 |
Hb_006810_030 |
0.1478232958 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |