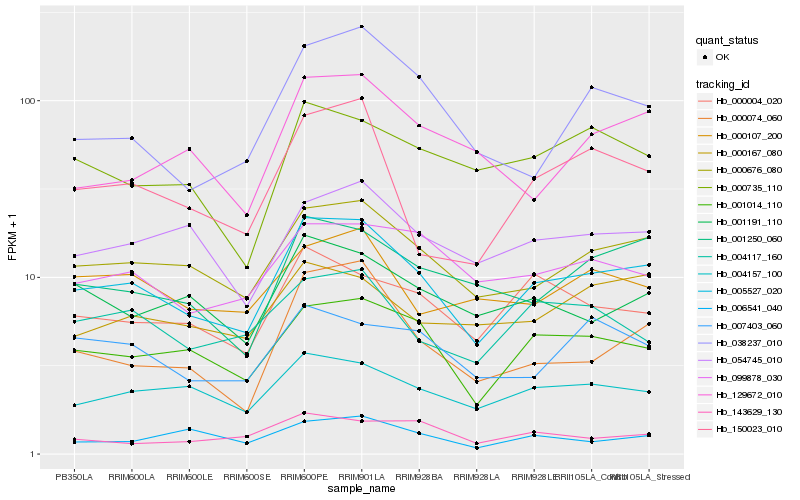
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005527_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 2 |
Hb_000676_080 |
0.0814321538 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 3 |
Hb_001014_110 |
0.0939952368 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 4 |
Hb_004157_100 |
0.1082564303 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 5 |
Hb_001250_060 |
0.1166112349 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 6 |
Hb_000004_020 |
0.1174555774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_054745_010 |
0.1209579022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001191_110 |
0.1222295769 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 9 |
Hb_099878_030 |
0.1230193006 |
- |
- |
PREDICTED: coiled-coil domain-containing protein R3HCC1L isoform X4 [Jatropha curcas] |
| 10 |
Hb_004117_160 |
0.1252625794 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |
| 11 |
Hb_000074_060 |
0.1253546583 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 12 |
Hb_150023_010 |
0.1267650174 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 13 |
Hb_000735_110 |
0.1280924749 |
- |
- |
fk506-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000167_080 |
0.1305081447 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 15 |
Hb_007403_060 |
0.131151301 |
- |
- |
PREDICTED: CAAX prenyl protease 2 [Jatropha curcas] |
| 16 |
Hb_000107_200 |
0.1321834419 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |
| 17 |
Hb_038237_010 |
0.1328068574 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 18 |
Hb_129672_010 |
0.1398025946 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 19 |
Hb_006541_040 |
0.1399589319 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 20 |
Hb_143629_130 |
0.1410500388 |
- |
- |
unnamed protein product [Coffea canephora] |