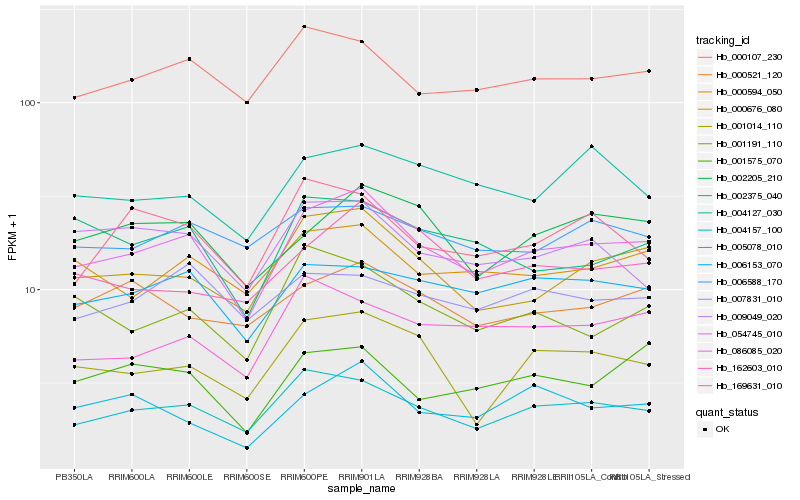
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054745_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004157_100 |
0.0736295225 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 3 |
Hb_000676_080 |
0.0821386543 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 4 |
Hb_000107_230 |
0.093693341 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_006153_070 |
0.0943058574 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000594_050 |
0.0967110541 |
- |
- |
PREDICTED: pH-response regulator protein palA/RIM20 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002205_210 |
0.100243945 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001014_110 |
0.1068322193 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 9 |
Hb_000521_120 |
0.1099839783 |
- |
- |
PREDICTED: cyclic pyranopterin monophosphate synthase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_006588_170 |
0.1100685702 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 11 |
Hb_001191_110 |
0.1102228302 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 12 |
Hb_002375_040 |
0.1102713207 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 13 |
Hb_086085_020 |
0.11081518 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 14 |
Hb_162603_010 |
0.1111914974 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_004127_030 |
0.1118706889 |
- |
- |
putative Rab geranylgeranyl transferase type II beta subunit family protein [Populus trichocarpa] |
| 16 |
Hb_005078_010 |
0.1119208643 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 17 |
Hb_009049_020 |
0.1122132715 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 18 |
Hb_169631_010 |
0.1123638087 |
- |
- |
MADS-box transcription factor, putative [Ricinus communis] |
| 19 |
Hb_007831_010 |
0.1124823653 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 20 |
Hb_001575_070 |
0.1133412332 |
- |
- |
PREDICTED: uncharacterized protein LOC105639885 [Jatropha curcas] |