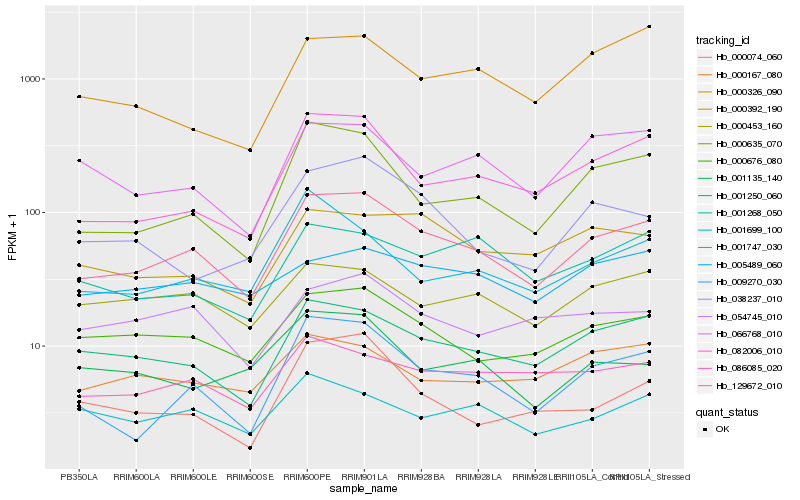
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129672_010 |
0.0 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 2 |
Hb_001250_060 |
0.0936736925 |
- |
- |
PREDICTED: signal recognition particle receptor subunit beta-like [Jatropha curcas] |
| 3 |
Hb_000676_080 |
0.1012441064 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C17D11.16 [Jatropha curcas] |
| 4 |
Hb_000635_070 |
0.105183049 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 5 |
Hb_009270_030 |
0.1059762994 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 6 |
Hb_082006_010 |
0.1074427143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_086085_020 |
0.1237855197 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 8 |
Hb_000392_190 |
0.1259215901 |
- |
- |
PREDICTED: NAP1-related protein 2 [Jatropha curcas] |
| 9 |
Hb_000453_160 |
0.1272442246 |
- |
- |
vacuolar protein sorting 26, vps26, putative [Ricinus communis] |
| 10 |
Hb_066768_010 |
0.1279594381 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_001268_050 |
0.1281662592 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 12 |
Hb_038237_010 |
0.1291994 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 13 |
Hb_054745_010 |
0.1309176924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000167_080 |
0.1331887451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 15 |
Hb_001699_100 |
0.1333416276 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 16 |
Hb_001747_030 |
0.1351781883 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 17 |
Hb_001135_140 |
0.1372678432 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 18 |
Hb_005489_060 |
0.1381955347 |
- |
- |
Anamorsin, putative [Ricinus communis] |
| 19 |
Hb_000074_060 |
0.1394463031 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 20 |
Hb_000326_090 |
0.1396063635 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |