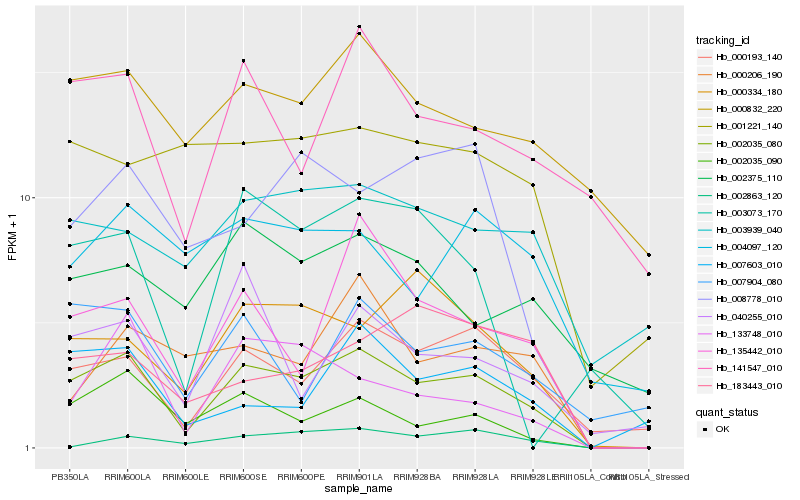
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002035_080 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000193_140 |
0.1733997524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_135442_010 |
0.1799948465 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 4 |
Hb_002035_090 |
0.1841968618 |
- |
- |
- |
| 5 |
Hb_000334_180 |
0.1922444762 |
- |
- |
hypothetical protein JCGZ_16779 [Jatropha curcas] |
| 6 |
Hb_133748_010 |
0.2080995768 |
- |
- |
- |
| 7 |
Hb_008778_010 |
0.2083774145 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_141547_010 |
0.2098730443 |
- |
- |
PREDICTED: ARF guanine-nucleotide exchange factor GNOM [Jatropha curcas] |
| 9 |
Hb_000206_190 |
0.2149400837 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 10 |
Hb_007603_010 |
0.2149799836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000832_220 |
0.2176364426 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X1 [Jatropha curcas] |
| 12 |
Hb_040255_010 |
0.2181710096 |
- |
- |
hypothetical protein JCGZ_08656 [Jatropha curcas] |
| 13 |
Hb_007904_080 |
0.2186553425 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 14 |
Hb_003939_040 |
0.2199155513 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 15 |
Hb_004097_120 |
0.2215873327 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_002375_110 |
0.2242663405 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_183443_010 |
0.2254668128 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 18 |
Hb_001221_140 |
0.2265478929 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_002863_120 |
0.2278382362 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 20 |
Hb_003073_170 |
0.2300127321 |
- |
- |
- |