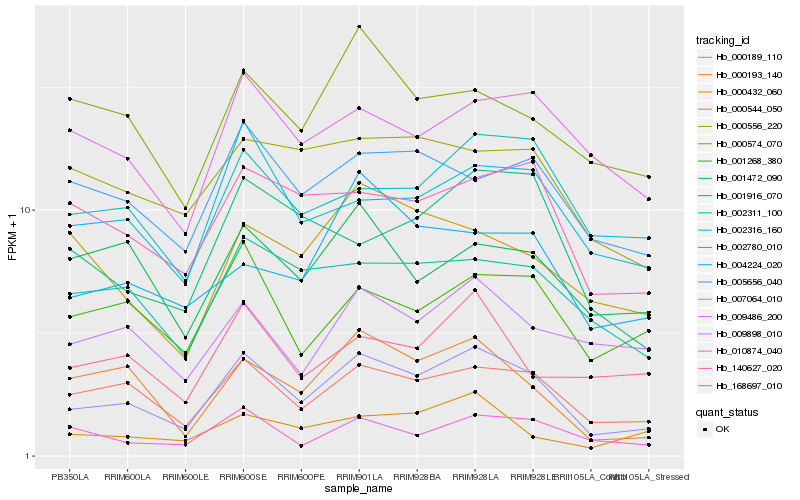
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007064_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 2 |
Hb_000189_110 |
0.1018449093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04750, mitochondrial [Jatropha curcas] |
| 3 |
Hb_000193_140 |
0.1295011761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002780_010 |
0.1396860712 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 5 |
Hb_140627_020 |
0.1416010778 |
- |
- |
PREDICTED: uncharacterized protein LOC105637253 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002311_100 |
0.142157421 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Jatropha curcas] |
| 7 |
Hb_002316_160 |
0.1440539199 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 8 |
Hb_010874_040 |
0.1475671734 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At3g13880 [Jatropha curcas] |
| 9 |
Hb_001916_070 |
0.1482133497 |
- |
- |
PREDICTED: phytochrome C [Jatropha curcas] |
| 10 |
Hb_001268_380 |
0.1488679053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g14080-like [Jatropha curcas] |
| 11 |
Hb_005656_040 |
0.1503920027 |
desease resistance |
Gene Name: T2SE |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_004224_020 |
0.1511809084 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 13 |
Hb_000432_060 |
0.1517385324 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000574_070 |
0.1538273712 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 35 [Jatropha curcas] |
| 15 |
Hb_009486_200 |
0.1542464302 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 16 |
Hb_009898_010 |
0.1551688773 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105645502 [Jatropha curcas] |
| 17 |
Hb_168697_010 |
0.1555092682 |
- |
- |
hypothetical protein JCGZ_16772 [Jatropha curcas] |
| 18 |
Hb_000556_220 |
0.1567015594 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 2-like [Jatropha curcas] |
| 19 |
Hb_000544_050 |
0.1568129544 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 20 |
Hb_001472_090 |
0.158587834 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 2 isoform X1 [Jatropha curcas] |