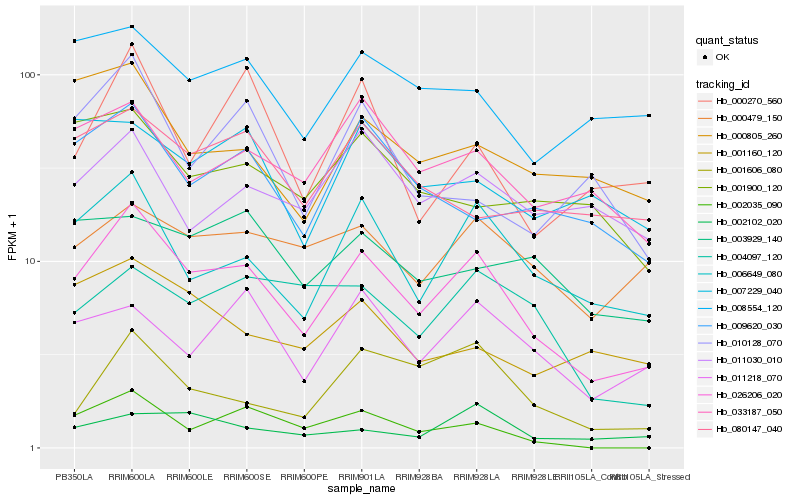
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026206_020 |
0.0 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006649_080 |
0.133008656 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 3 |
Hb_033187_050 |
0.1659642677 |
- |
- |
PREDICTED: exocyst complex component SEC15B [Jatropha curcas] |
| 4 |
Hb_009620_030 |
0.1730881569 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP59 [Jatropha curcas] |
| 5 |
Hb_000479_150 |
0.1740187988 |
- |
- |
glycosyl transferase family 8 family protein [Populus trichocarpa] |
| 6 |
Hb_011218_070 |
0.1754036671 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 7 |
Hb_008554_120 |
0.1772574216 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 8 |
Hb_000805_260 |
0.1776726792 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X1 [Jatropha curcas] |
| 9 |
Hb_080147_040 |
0.183440515 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 10 |
Hb_000270_560 |
0.1834419725 |
- |
- |
PREDICTED: U-box domain-containing protein 38-like [Jatropha curcas] |
| 11 |
Hb_001606_080 |
0.1835551627 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g04740 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004097_120 |
0.1849419705 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001160_120 |
0.1850968517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002102_020 |
0.18766889 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 15 |
Hb_011030_010 |
0.1881163327 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_010128_070 |
0.1881946725 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 17 |
Hb_003929_140 |
0.1886368173 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001900_120 |
0.1894828074 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 19 |
Hb_007229_040 |
0.1910190292 |
- |
- |
PREDICTED: protein ARABIDILLO 1-like [Jatropha curcas] |
| 20 |
Hb_002035_090 |
0.1920434388 |
- |
- |
- |