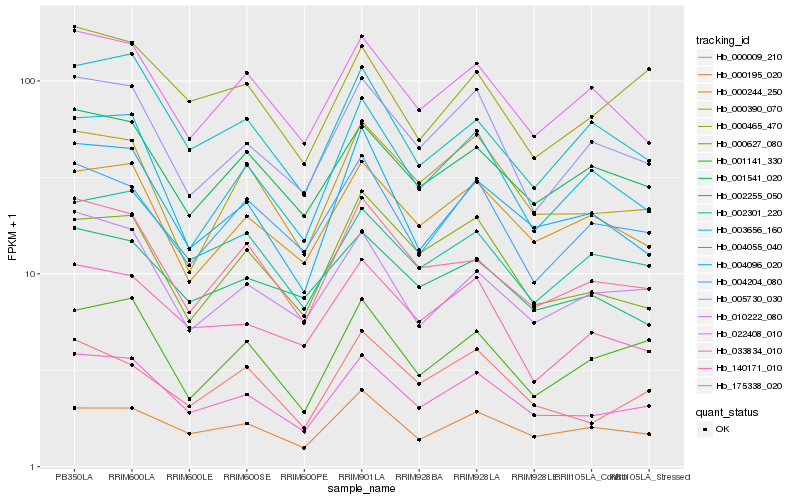
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033834_010 |
0.0 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 2 |
Hb_005730_030 |
0.0764865653 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 3 |
Hb_175338_020 |
0.0800146095 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 4 |
Hb_004096_020 |
0.0805212443 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like, partial [Fragaria vesca subsp. vesca] |
| 5 |
Hb_001541_020 |
0.0805251579 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 6 |
Hb_000195_020 |
0.080859653 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19191, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002301_220 |
0.0816269702 |
transcription factor |
TF Family: C3H |
PREDICTED: protein FRIGIDA-ESSENTIAL 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_022408_010 |
0.0830753656 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 9 |
Hb_000244_250 |
0.0831114094 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010222_080 |
0.0831331874 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 11 |
Hb_000390_070 |
0.0848128176 |
- |
- |
PREDICTED: uncharacterized protein LOC105632478 isoform X1 [Jatropha curcas] |
| 12 |
Hb_140171_010 |
0.0871652985 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 13 |
Hb_004055_040 |
0.0877509753 |
- |
- |
Phospholipid-transporting ATPase, putative [Ricinus communis] |
| 14 |
Hb_000009_210 |
0.0881745335 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g01390 [Jatropha curcas] |
| 15 |
Hb_000627_080 |
0.0903355335 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 16 |
Hb_001141_330 |
0.0908894434 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14850 [Jatropha curcas] |
| 17 |
Hb_002255_050 |
0.0910121455 |
- |
- |
PREDICTED: probable isoleucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_003656_160 |
0.0918162999 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein B456_013G226400 [Gossypium raimondii] |
| 19 |
Hb_004204_080 |
0.0931040552 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 20 |
Hb_000465_470 |
0.0934954882 |
- |
- |
unknown [Picea sitchensis] |