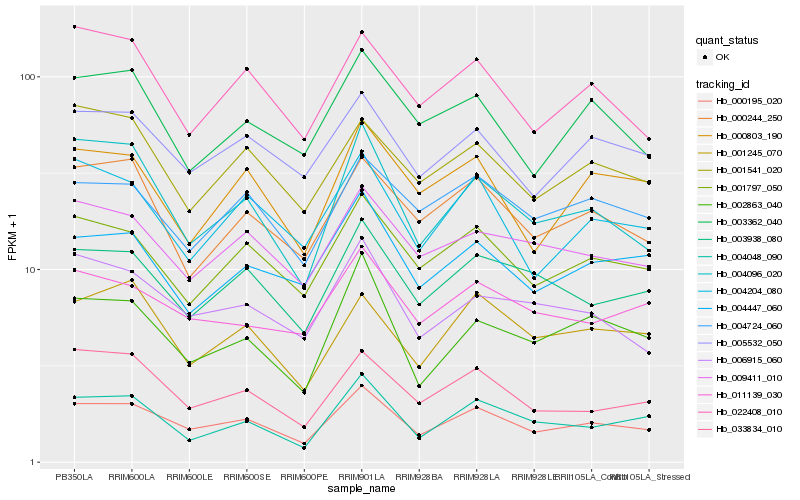
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000195_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19191, mitochondrial [Jatropha curcas] |
| 2 |
Hb_001797_050 |
0.069256198 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004096_020 |
0.0761517277 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like, partial [Fragaria vesca subsp. vesca] |
| 4 |
Hb_005532_050 |
0.0770193386 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 5 |
Hb_003938_080 |
0.0776699412 |
- |
- |
PREDICTED: uncharacterized protein LOC105644554 [Jatropha curcas] |
| 6 |
Hb_033834_010 |
0.080859653 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 7 |
Hb_009411_010 |
0.0823629109 |
- |
- |
PREDICTED: uncharacterized protein LOC105645272 [Jatropha curcas] |
| 8 |
Hb_004204_080 |
0.0824528039 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 9 |
Hb_002863_040 |
0.0850588281 |
- |
- |
PREDICTED: uncharacterized protein LOC105630268 [Jatropha curcas] |
| 10 |
Hb_022408_010 |
0.0858289824 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 11 |
Hb_000244_250 |
0.0862440534 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006915_060 |
0.0866418872 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 13 |
Hb_004724_060 |
0.0875939004 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001541_020 |
0.0895040319 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 15 |
Hb_004447_060 |
0.0903349029 |
- |
- |
PREDICTED: uncharacterized protein LOC105636329 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003362_040 |
0.090475719 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 17 |
Hb_001245_070 |
0.0905120271 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 18 |
Hb_000803_190 |
0.0905925237 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011139_030 |
0.0906713114 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 20 |
Hb_004048_090 |
0.0908098355 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |