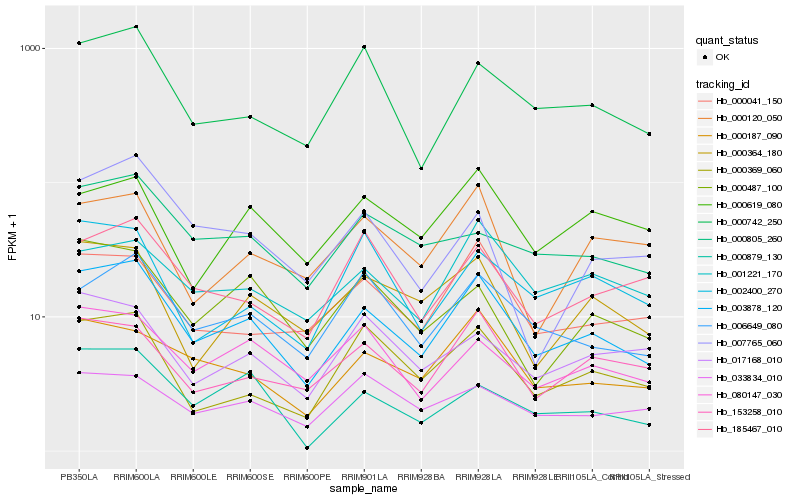
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003878_120 |
0.0 |
transcription factor |
TF Family: MBF1 |
PREDICTED: multiprotein-bridging factor 1b-like [Elaeis guineensis] |
| 2 |
Hb_000187_090 |
0.1226828946 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 3 |
Hb_000742_250 |
0.1314317341 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 4 |
Hb_006649_080 |
0.1316220825 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 5 |
Hb_000041_150 |
0.133147251 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 6 |
Hb_000364_180 |
0.1344198555 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130 [Jatropha curcas] |
| 7 |
Hb_153258_010 |
0.1351614475 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 8 |
Hb_080147_030 |
0.1397969183 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000369_060 |
0.1411162887 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 10 |
Hb_001221_170 |
0.1441261982 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000487_100 |
0.1456633629 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 12 |
Hb_000805_260 |
0.1463364541 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000879_130 |
0.1463374213 |
- |
- |
- |
| 14 |
Hb_185467_010 |
0.1544157394 |
- |
- |
- |
| 15 |
Hb_033834_010 |
0.1551195408 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_002400_270 |
0.1568394733 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_017168_010 |
0.1568616488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000120_050 |
0.1569550909 |
- |
- |
PREDICTED: protein ELF4-LIKE 1 [Jatropha curcas] |
| 19 |
Hb_000619_080 |
0.1575186058 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 21 [Jatropha curcas] |
| 20 |
Hb_007765_060 |
0.1591649964 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |