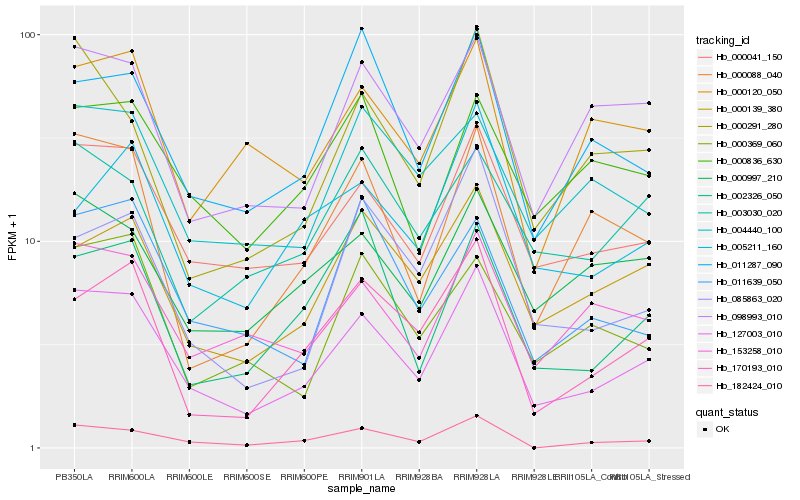
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_127003_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 2 |
Hb_000041_150 |
0.0864556591 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 3 |
Hb_182424_010 |
0.1263794529 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 4 |
Hb_170193_010 |
0.1274808078 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 5 |
Hb_153258_010 |
0.1318852047 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 6 |
Hb_000139_380 |
0.1319003642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_098993_010 |
0.1335811112 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000088_040 |
0.134037962 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 9 |
Hb_000997_210 |
0.1405593451 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 10 |
Hb_000291_280 |
0.1407883042 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 11 |
Hb_011287_090 |
0.1468575344 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 12 |
Hb_085863_020 |
0.1486677272 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 13 |
Hb_000120_050 |
0.1503426532 |
- |
- |
PREDICTED: protein ELF4-LIKE 1 [Jatropha curcas] |
| 14 |
Hb_011639_050 |
0.1503691172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004440_100 |
0.1510710332 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 16 |
Hb_005211_160 |
0.1522451086 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 17 |
Hb_000369_060 |
0.1524254465 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_000836_630 |
0.1538174481 |
- |
- |
PREDICTED: WD repeat-containing protein 44 [Jatropha curcas] |
| 19 |
Hb_003030_020 |
0.1553252346 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |
| 20 |
Hb_002326_050 |
0.1558729965 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |