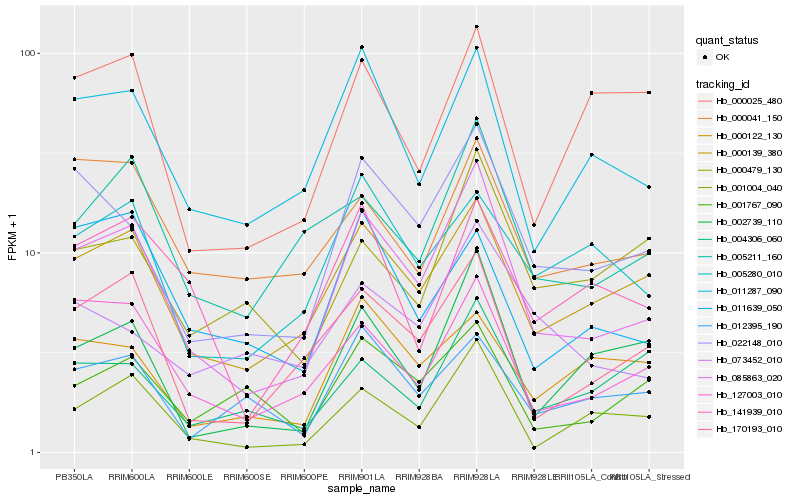
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085863_020 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 2 |
Hb_022148_010 |
0.1330439997 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 3 |
Hb_000139_380 |
0.1380902109 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_170193_010 |
0.1454750474 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 5 |
Hb_127003_010 |
0.1486677272 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 6 |
Hb_001004_040 |
0.1521793377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011287_090 |
0.1545043257 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 8 |
Hb_005211_160 |
0.1633532281 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 9 |
Hb_002739_110 |
0.1641586012 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 10 |
Hb_000041_150 |
0.1850185866 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 11 |
Hb_141939_010 |
0.1864040582 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 3, chloroplastic-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001767_090 |
0.1868443301 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 13 |
Hb_012395_190 |
0.187515353 |
- |
- |
hypothetical protein B456_005G053800 [Gossypium raimondii] |
| 14 |
Hb_005280_010 |
0.1898441145 |
- |
- |
PREDICTED: protein phosphatase 2C 29 [Jatropha curcas] |
| 15 |
Hb_073452_010 |
0.1908018864 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 16 |
Hb_000479_130 |
0.1912465147 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350 [Jatropha curcas] |
| 17 |
Hb_004306_060 |
0.1934726463 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000122_130 |
0.1952113538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000025_480 |
0.1953003998 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 20 |
Hb_011639_050 |
0.1961009371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |