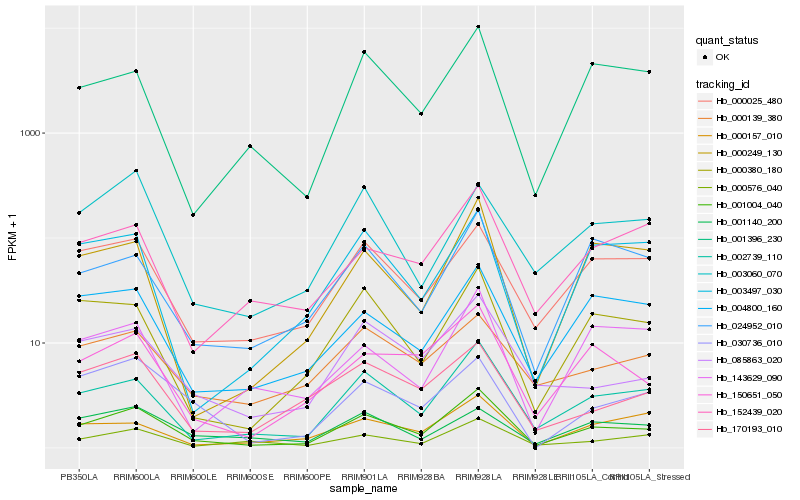
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001004_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002739_110 |
0.1107394897 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 3 |
Hb_150651_050 |
0.1458781895 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 4 |
Hb_000380_180 |
0.1487931378 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_085863_020 |
0.1521793377 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 6 |
Hb_000025_480 |
0.1527553052 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000249_130 |
0.1580606134 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 8 |
Hb_024952_010 |
0.1627418245 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 9 |
Hb_004800_160 |
0.1635265076 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 10 |
Hb_001396_230 |
0.1639350057 |
- |
- |
hypothetical protein MIMGU_mgv1a023992mg, partial [Erythranthe guttata] |
| 11 |
Hb_000576_040 |
0.1646167814 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15130 [Jatropha curcas] |
| 12 |
Hb_170193_010 |
0.167817872 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 13 |
Hb_003497_030 |
0.16959619 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 14 |
Hb_152439_020 |
0.17010879 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_030736_010 |
0.1714812722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000139_380 |
0.1765058509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_143629_090 |
0.1785753362 |
- |
- |
PREDICTED: uncharacterized protein LOC105647358 [Jatropha curcas] |
| 18 |
Hb_003060_070 |
0.1819517624 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001140_200 |
0.1829441291 |
- |
- |
PREDICTED: uncharacterized protein LOC105648845 [Jatropha curcas] |
| 20 |
Hb_000157_010 |
0.1834053458 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |