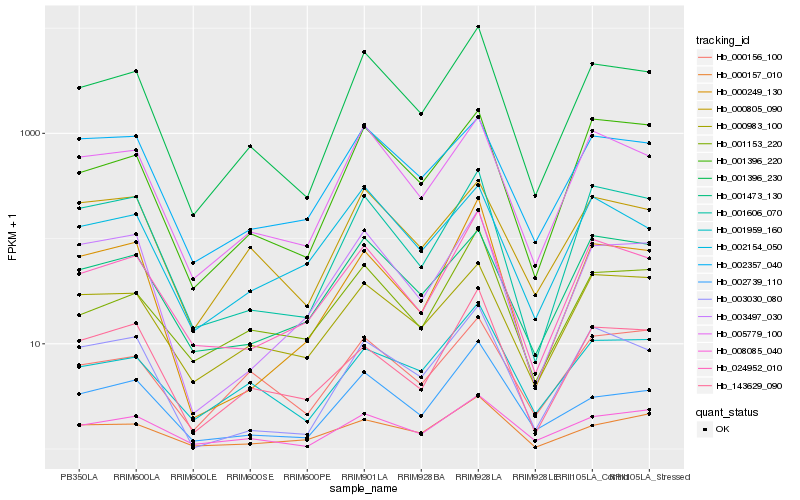
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001396_230 |
0.0 |
- |
- |
hypothetical protein MIMGU_mgv1a023992mg, partial [Erythranthe guttata] |
| 2 |
Hb_005779_100 |
0.0855659803 |
- |
- |
DNAJ [Theobroma cacao] |
| 3 |
Hb_024952_010 |
0.0981572411 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 4 |
Hb_008085_040 |
0.1027184249 |
- |
- |
Pentatricopeptide repeat-containing protein, mitochondrial [Glycine soja] |
| 5 |
Hb_002739_110 |
0.105436113 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 6 |
Hb_001606_070 |
0.1074350971 |
- |
- |
fimbrin, putative [Ricinus communis] |
| 7 |
Hb_003030_080 |
0.1123414771 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_001959_160 |
0.118440129 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 9 |
Hb_001396_220 |
0.1190833146 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 10 |
Hb_003497_030 |
0.1221984166 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 11 |
Hb_001153_220 |
0.1239385149 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Jatropha curcas] |
| 12 |
Hb_002357_040 |
0.126273124 |
- |
- |
latex-abundant protein [Hevea brasiliensis] |
| 13 |
Hb_000249_130 |
0.1279506057 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 14 |
Hb_000805_090 |
0.1300606821 |
- |
- |
hypothetical protein POPTR_0009s03890g [Populus trichocarpa] |
| 15 |
Hb_000157_010 |
0.1305991909 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 16 |
Hb_001473_130 |
0.1308014902 |
- |
- |
PREDICTED: ubiquinone biosynthesis monooxygenase COQ6 [Jatropha curcas] |
| 17 |
Hb_000983_100 |
0.1333157509 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X1 [Vitis vinifera] |
| 18 |
Hb_143629_090 |
0.135186982 |
- |
- |
PREDICTED: uncharacterized protein LOC105647358 [Jatropha curcas] |
| 19 |
Hb_002154_050 |
0.1365518038 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000156_100 |
0.139226402 |
- |
- |
PREDICTED: uncharacterized mitochondrial protein ymf11 [Jatropha curcas] |