| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
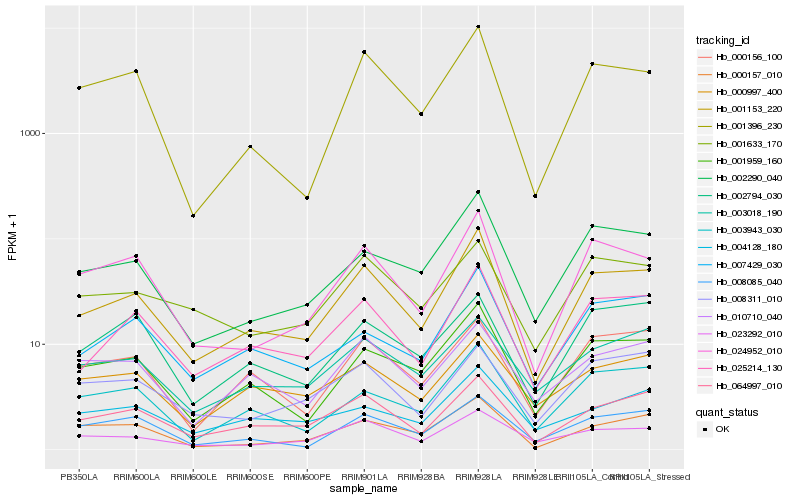
Hb_001153_220 |
0.0 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Jatropha curcas] |
| 2 |
Hb_025214_130 |
0.0877370496 |
- |
- |
PREDICTED: uncharacterized protein LOC105640854 [Jatropha curcas] |
| 3 |
Hb_064997_010 |
0.0984149674 |
- |
- |
hypothetical protein JCGZ_19852 [Jatropha curcas] |
| 4 |
Hb_001959_160 |
0.1016640371 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 5 |
Hb_024952_010 |
0.1073502474 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 6 |
Hb_002290_040 |
0.1100388009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_023292_010 |
0.1216986997 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 8 |
Hb_003943_030 |
0.1221829261 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g32415, mitochondrial [Jatropha curcas] |
| 9 |
Hb_007429_030 |
0.1225503883 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001396_230 |
0.1239385149 |
- |
- |
hypothetical protein MIMGU_mgv1a023992mg, partial [Erythranthe guttata] |
| 11 |
Hb_000157_010 |
0.1272151713 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 12 |
Hb_000156_100 |
0.1336228159 |
- |
- |
PREDICTED: uncharacterized mitochondrial protein ymf11 [Jatropha curcas] |
| 13 |
Hb_008085_040 |
0.1348062719 |
- |
- |
Pentatricopeptide repeat-containing protein, mitochondrial [Glycine soja] |
| 14 |
Hb_003018_190 |
0.1399396635 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g31070, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001633_170 |
0.1418965538 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000997_400 |
0.1467952054 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g50270 [Jatropha curcas] |
| 17 |
Hb_010710_040 |
0.1480034088 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_004128_180 |
0.1485334348 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_008311_010 |
0.1486555586 |
- |
- |
hypothetical protein CARUB_v10003781mg [Capsella rubella] |
| 20 |
Hb_002794_030 |
0.1486564292 |
- |
- |
hypothetical protein JCGZ_20185 [Jatropha curcas] |