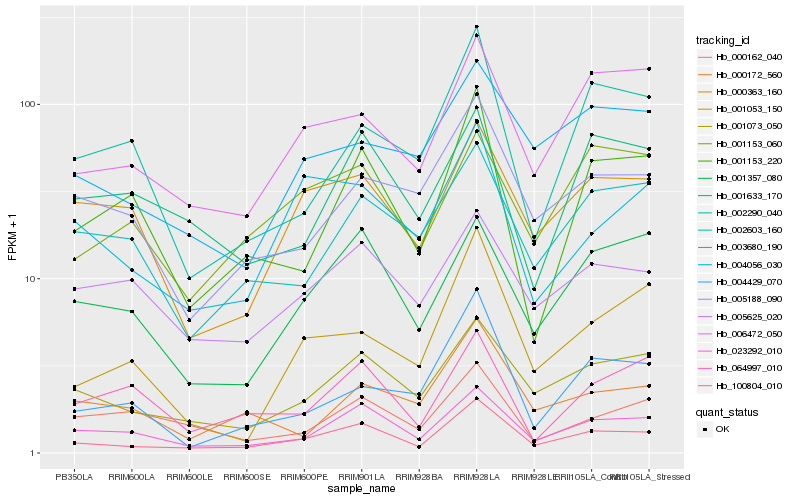
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_100804_010 |
0.0 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 2 |
Hb_023292_010 |
0.11635174 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_001073_050 |
0.1277583783 |
- |
- |
- |
| 4 |
Hb_006472_050 |
0.1514670521 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 7 isoform X2 [Elaeis guineensis] |
| 5 |
Hb_001153_220 |
0.1565124717 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Jatropha curcas] |
| 6 |
Hb_001357_080 |
0.1589156167 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 7 |
Hb_000363_160 |
0.1625441458 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 8 |
Hb_003680_190 |
0.1630489478 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 9 |
Hb_005188_090 |
0.1690656232 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 10 |
Hb_005625_020 |
0.1707493218 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004056_030 |
0.1754900399 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 12 |
Hb_001053_150 |
0.1757181002 |
- |
- |
- |
| 13 |
Hb_002603_160 |
0.1760256485 |
- |
- |
Arginyl-tRNA--protein transferase, putative [Ricinus communis] |
| 14 |
Hb_001153_060 |
0.1775623408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004429_070 |
0.1789176067 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 16 |
Hb_000172_560 |
0.1809527452 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 17 |
Hb_064997_010 |
0.181078676 |
- |
- |
hypothetical protein JCGZ_19852 [Jatropha curcas] |
| 18 |
Hb_001633_170 |
0.1815626993 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002290_040 |
0.1816431729 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000162_040 |
0.1820206606 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |