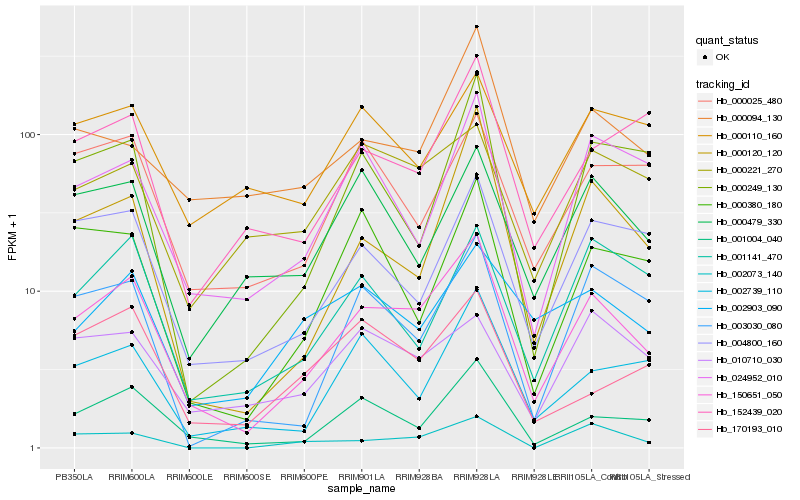
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150651_050 |
0.0 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 2 |
Hb_001004_040 |
0.1458781895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004800_160 |
0.172233999 |
- |
- |
Uncharacterized protein TCM_000403 [Theobroma cacao] |
| 4 |
Hb_000120_120 |
0.1806128634 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2 [Jatropha curcas] |
| 5 |
Hb_003030_080 |
0.1815895409 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 6 |
Hb_002739_110 |
0.1828666595 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_024952_010 |
0.1844034114 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 8 |
Hb_000249_130 |
0.1891644395 |
- |
- |
hypothetical protein RCOM_0254640 [Ricinus communis] |
| 9 |
Hb_000479_330 |
0.1892217166 |
- |
- |
PREDICTED: uncharacterized protein LOC105644113 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000025_480 |
0.1892510421 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 11 |
Hb_152439_020 |
0.1899179791 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001141_470 |
0.1904534945 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 13 |
Hb_002073_140 |
0.1908365031 |
- |
- |
- |
| 14 |
Hb_002903_090 |
0.1913562219 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 15 |
Hb_000094_130 |
0.1921816717 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 16 |
Hb_000221_270 |
0.1932173447 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 17 |
Hb_000380_180 |
0.1941612365 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_010710_030 |
0.1972810685 |
transcription factor |
TF Family: G2-like |
PREDICTED: transcription factor LUX [Jatropha curcas] |
| 19 |
Hb_000110_160 |
0.1978334498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_170193_010 |
0.1978337089 |
- |
- |
JHL25H03.10 [Jatropha curcas] |