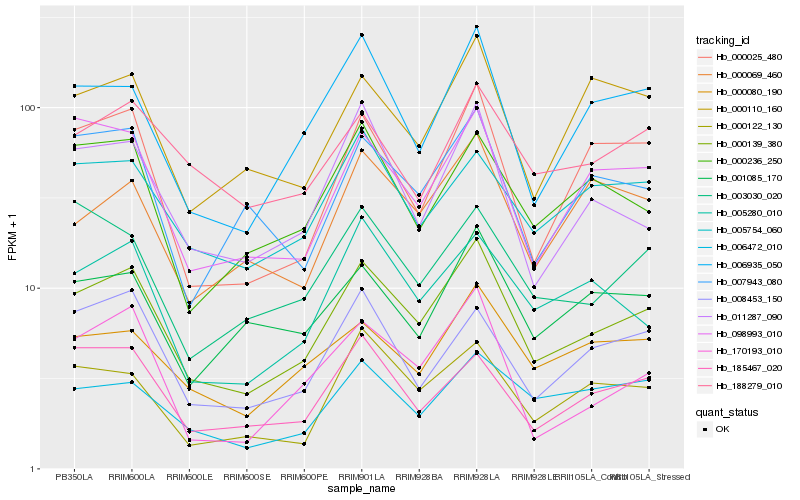
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_380 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_006935_050 |
0.1106225027 |
- |
- |
PREDICTED: probable glutathione peroxidase 2 [Populus euphratica] |
| 3 |
Hb_098993_010 |
0.1119323045 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000025_480 |
0.1149064541 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein 10 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000080_190 |
0.1162989791 |
- |
- |
- |
| 6 |
Hb_011287_090 |
0.117233477 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 7 |
Hb_005280_010 |
0.1202103664 |
- |
- |
PREDICTED: protein phosphatase 2C 29 [Jatropha curcas] |
| 8 |
Hb_170193_010 |
0.1207181451 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 9 |
Hb_185467_020 |
0.1221290846 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 10 |
Hb_008453_150 |
0.1225675471 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005754_060 |
0.1230433246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000236_250 |
0.1235062299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000122_130 |
0.1243004935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001085_170 |
0.1243982989 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007943_080 |
0.1264906 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006472_010 |
0.1279788705 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 17 |
Hb_000110_160 |
0.1280280053 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_188279_010 |
0.129100019 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC9-like [Tarenaya hassleriana] |
| 19 |
Hb_000069_460 |
0.1291049733 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 20 |
Hb_003030_020 |
0.1292931855 |
- |
- |
PREDICTED: uncharacterized protein LOC105632544 [Jatropha curcas] |