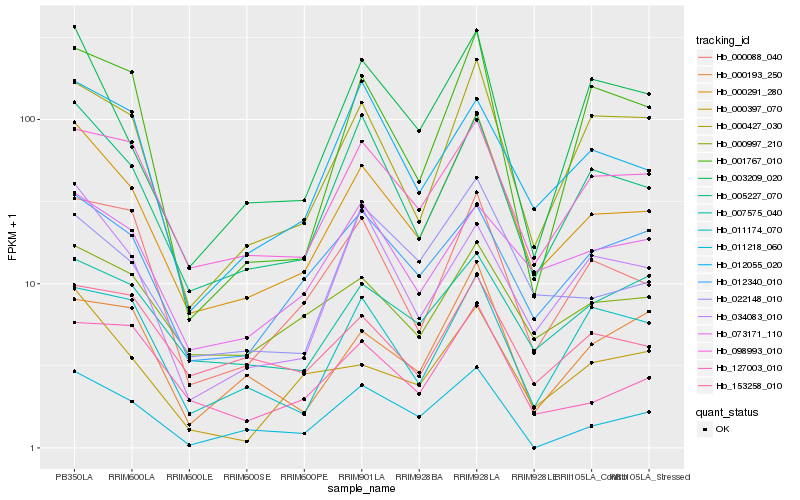
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_280 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 2 |
Hb_005227_070 |
0.1102120437 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_000427_030 |
0.1290046691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_098993_010 |
0.130867147 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000088_040 |
0.1316723815 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_022148_010 |
0.1335015228 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 7 |
Hb_127003_010 |
0.1407883042 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 8 |
Hb_000997_210 |
0.1409177733 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 9 |
Hb_003209_020 |
0.1424215678 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 10 |
Hb_000397_070 |
0.1460586026 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 11 |
Hb_012340_010 |
0.1485189496 |
- |
- |
PREDICTED: B-cell receptor-associated protein 31 [Jatropha curcas] |
| 12 |
Hb_011218_060 |
0.1500001586 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 13 |
Hb_012055_020 |
0.1505237966 |
- |
- |
PREDICTED: metal transporter Nramp3 [Jatropha curcas] |
| 14 |
Hb_034083_010 |
0.1524046803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_073171_110 |
0.1535722338 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 16 |
Hb_153258_010 |
0.1539785679 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 17 |
Hb_001767_010 |
0.1568442605 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 18 |
Hb_000193_250 |
0.1581773487 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 19 |
Hb_011174_070 |
0.1583299356 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g06540-like [Jatropha curcas] |
| 20 |
Hb_007575_040 |
0.1583369077 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor VIP1-like [Jatropha curcas] |