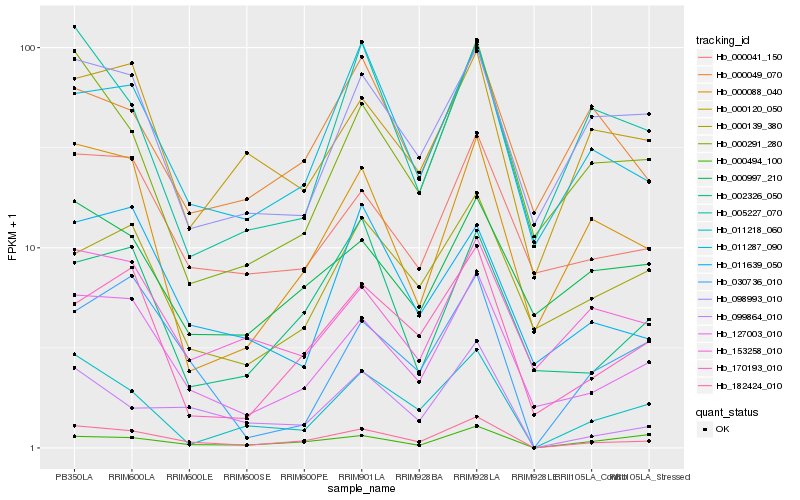
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_182424_010 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 2 |
Hb_127003_010 |
0.1263794529 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 3 |
Hb_099864_010 |
0.1395756486 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 4 |
Hb_011287_090 |
0.1563657566 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 5 |
Hb_011218_060 |
0.1566303909 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 6 |
Hb_170193_010 |
0.1620548891 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 7 |
Hb_000041_150 |
0.1686231962 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 8 |
Hb_000494_100 |
0.1698401181 |
- |
- |
H(\+)-transporting atpase plant/fungi plasma membrane type, putative [Ricinus communis] |
| 9 |
Hb_000088_040 |
0.1727415701 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 10 |
Hb_000291_280 |
0.1772994025 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 11 |
Hb_000120_050 |
0.1811836148 |
- |
- |
PREDICTED: protein ELF4-LIKE 1 [Jatropha curcas] |
| 12 |
Hb_030736_010 |
0.1828213148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_153258_010 |
0.1846600181 |
- |
- |
PREDICTED: uncharacterized protein LOC105642325 [Jatropha curcas] |
| 14 |
Hb_098993_010 |
0.1850016009 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002326_050 |
0.1874200456 |
- |
- |
PREDICTED: uncharacterized protein LOC105633701 [Jatropha curcas] |
| 16 |
Hb_000139_380 |
0.1919084916 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_011639_050 |
0.1931714343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000997_210 |
0.1935914958 |
- |
- |
PREDICTED: protein transport protein sec23-1 [Jatropha curcas] |
| 19 |
Hb_000049_070 |
0.194341473 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005227_070 |
0.1965749494 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |