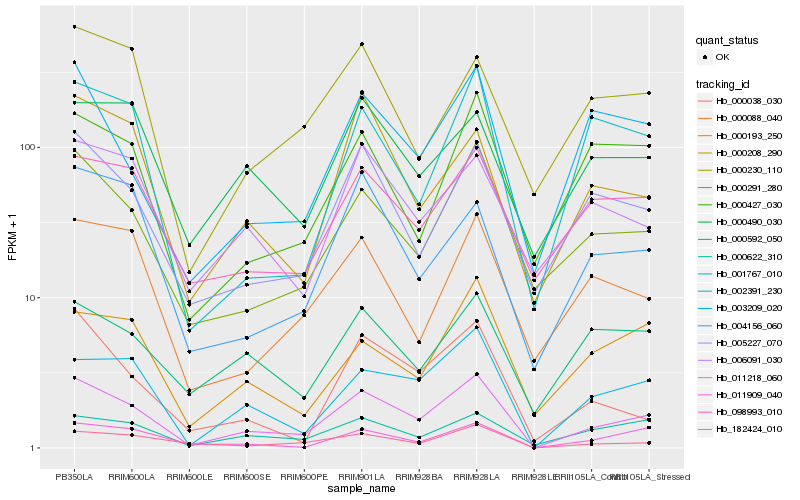
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011218_060 |
0.0 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 2 |
Hb_002391_230 |
0.1382822963 |
- |
- |
hypothetical protein RCOM_0745080 [Ricinus communis] |
| 3 |
Hb_000291_280 |
0.1500001586 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 4 |
Hb_182424_010 |
0.1566303909 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 5 |
Hb_005227_070 |
0.160351207 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000622_310 |
0.1685334882 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 7 |
Hb_011909_040 |
0.1745838938 |
- |
- |
hypothetical protein JCGZ_00750 [Jatropha curcas] |
| 8 |
Hb_003209_020 |
0.1752032914 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 9 |
Hb_098993_010 |
0.1753852109 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000038_030 |
0.1759613451 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 11 |
Hb_000208_290 |
0.1787647199 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004156_060 |
0.1788344458 |
- |
- |
neutral/alkaline invertase 1 [Hevea brasiliensis] |
| 13 |
Hb_000088_040 |
0.1795041134 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 14 |
Hb_001767_010 |
0.1802272996 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 15 |
Hb_000193_250 |
0.1811684417 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 16 |
Hb_006091_030 |
0.1837261024 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000230_110 |
0.184813696 |
- |
- |
PREDICTED: uncharacterized protein LOC105647634 [Jatropha curcas] |
| 18 |
Hb_000592_050 |
0.185346847 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 19 |
Hb_000427_030 |
0.1853958779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000490_030 |
0.1872059367 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |