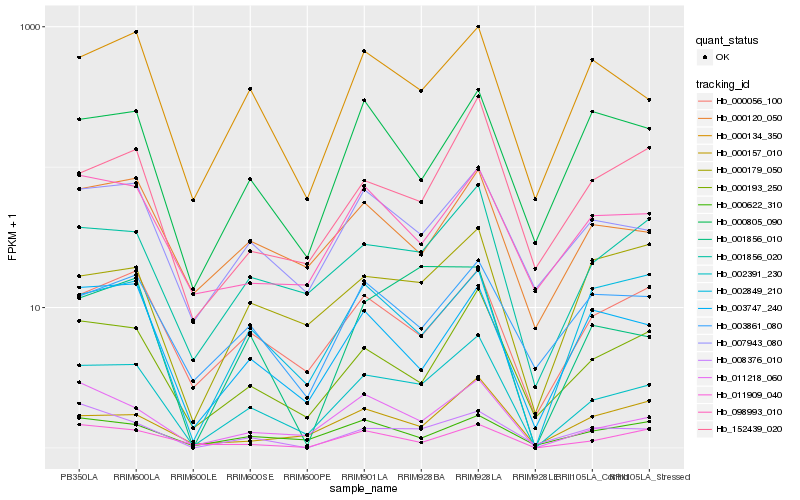
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_230 |
0.0 |
- |
- |
hypothetical protein RCOM_0745080 [Ricinus communis] |
| 2 |
Hb_011218_060 |
0.1382822963 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 3 |
Hb_001856_020 |
0.1386965045 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 4 |
Hb_000193_250 |
0.1442879375 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61400 [Jatropha curcas] |
| 5 |
Hb_000056_100 |
0.1649112448 |
- |
- |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000134_350 |
0.1654095645 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 8-like [Jatropha curcas] |
| 7 |
Hb_007943_080 |
0.1662582485 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003747_240 |
0.1716884528 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002849_210 |
0.1741224388 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001856_010 |
0.175338882 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_152439_020 |
0.1761105203 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000622_310 |
0.1807482713 |
- |
- |
PREDICTED: BON1-associated protein 2 [Jatropha curcas] |
| 13 |
Hb_000120_050 |
0.1815676956 |
- |
- |
PREDICTED: protein ELF4-LIKE 1 [Jatropha curcas] |
| 14 |
Hb_000179_050 |
0.1826473446 |
- |
- |
40S ribosomal protein S17A [Hevea brasiliensis] |
| 15 |
Hb_000157_010 |
0.1845207043 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 16 |
Hb_008376_010 |
0.1854590798 |
- |
- |
- |
| 17 |
Hb_011909_040 |
0.1876063947 |
- |
- |
hypothetical protein JCGZ_00750 [Jatropha curcas] |
| 18 |
Hb_000805_090 |
0.1880696765 |
- |
- |
hypothetical protein POPTR_0009s03890g [Populus trichocarpa] |
| 19 |
Hb_003861_080 |
0.1882427816 |
- |
- |
PREDICTED: DNA polymerase kappa isoform X2 [Jatropha curcas] |
| 20 |
Hb_098993_010 |
0.1893209775 |
- |
- |
PREDICTED: chaperone protein dnaJ 72 isoform X2 [Jatropha curcas] |