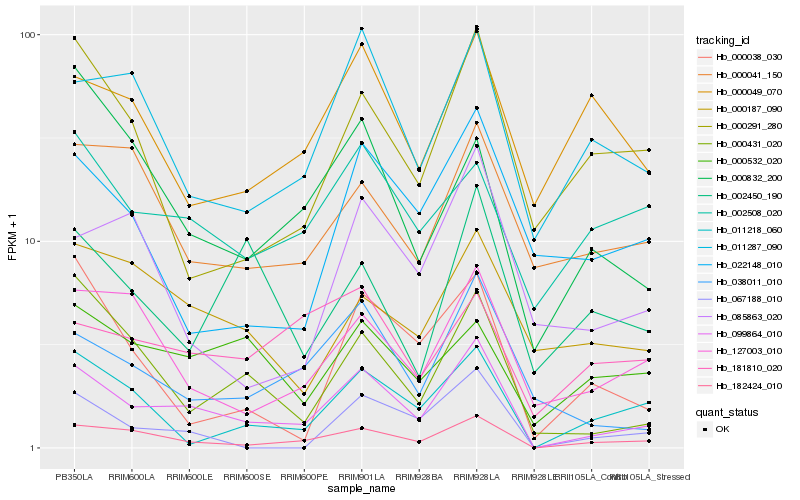
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_099864_010 |
0.0 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 2 |
Hb_182424_010 |
0.1395756486 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 3 |
Hb_038011_010 |
0.1951176284 |
- |
- |
hypothetical protein POPTR_0031s004401g, partial [Populus trichocarpa] |
| 4 |
Hb_000431_020 |
0.2045634003 |
- |
- |
hypothetical protein JCGZ_06801 [Jatropha curcas] |
| 5 |
Hb_011218_060 |
0.2116914032 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 6 |
Hb_127003_010 |
0.2119252434 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 7 |
Hb_011287_090 |
0.2140724427 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 8 |
Hb_067188_010 |
0.2233097955 |
- |
- |
- |
| 9 |
Hb_000187_090 |
0.2312880245 |
- |
- |
PREDICTED: magnesium transporter MRS2-3 [Jatropha curcas] |
| 10 |
Hb_002508_020 |
0.2320302452 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 11 |
Hb_000532_020 |
0.2321040399 |
- |
- |
beta-tubulin [Bouteloua eriopoda] |
| 12 |
Hb_000291_280 |
0.2321965145 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: myosin-11 [Jatropha curcas] |
| 13 |
Hb_000041_150 |
0.2326880424 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 14 |
Hb_000832_200 |
0.234216201 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000038_030 |
0.2372914542 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 16 |
Hb_022148_010 |
0.238118009 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 17 |
Hb_085863_020 |
0.2392183462 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 18 |
Hb_181810_020 |
0.2423979655 |
- |
- |
- |
| 19 |
Hb_002450_190 |
0.2448581441 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 20 |
Hb_000049_070 |
0.2449825558 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |