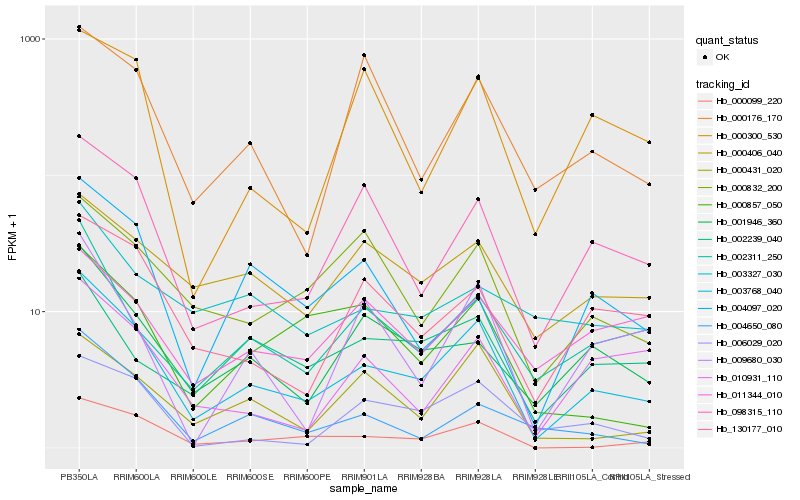
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003768_040 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_000099_220 |
0.1547759712 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_098315_110 |
0.1699330297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002311_250 |
0.1729708733 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 5 |
Hb_001946_360 |
0.1736363437 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 6 |
Hb_130177_010 |
0.1793452633 |
- |
- |
hypothetical protein RCOM_0897890 [Ricinus communis] |
| 7 |
Hb_000431_020 |
0.1884274822 |
- |
- |
hypothetical protein JCGZ_06801 [Jatropha curcas] |
| 8 |
Hb_006029_020 |
0.1928963486 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1, partial [Vitis vinifera] |
| 9 |
Hb_000832_200 |
0.1955521831 |
- |
- |
2-oxoglutarate dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_010931_110 |
0.1972520719 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_000406_040 |
0.1980401412 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 12 |
Hb_004650_080 |
0.2010236714 |
- |
- |
- |
| 13 |
Hb_000857_050 |
0.2026953848 |
- |
- |
PREDICTED: protein YLS9 [Jatropha curcas] |
| 14 |
Hb_009680_030 |
0.2028696618 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 15 |
Hb_011344_010 |
0.2056665412 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 16 |
Hb_000300_530 |
0.2066835784 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0286299-like [Gossypium raimondii] |
| 17 |
Hb_002239_040 |
0.2068670668 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 18 |
Hb_000176_170 |
0.2081100217 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 19 |
Hb_004097_020 |
0.2104331031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003327_030 |
0.2112476793 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |