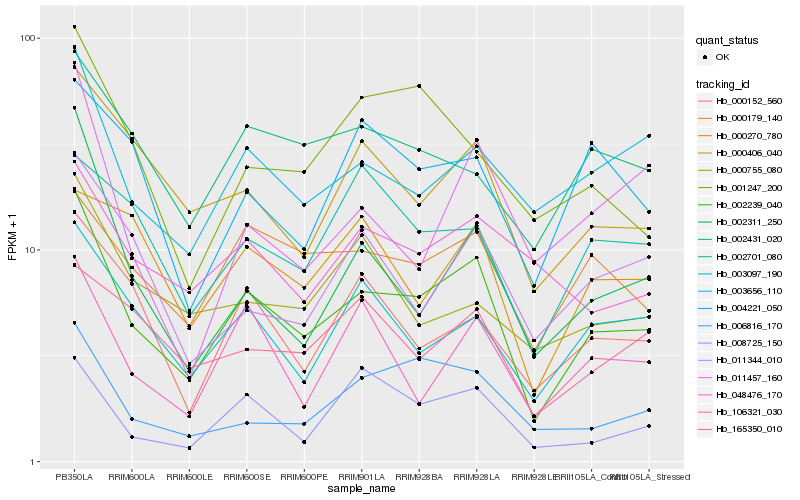
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002239_040 |
0.0 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_000406_040 |
0.152219038 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 3 |
Hb_003656_110 |
0.1539676563 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_003097_190 |
0.1559550133 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 5 |
Hb_002311_250 |
0.1611220352 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 6 |
Hb_006816_170 |
0.1637702384 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 7 |
Hb_000152_560 |
0.1660041879 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 8 |
Hb_000179_140 |
0.1664573081 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_011344_010 |
0.1702816356 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 10 |
Hb_002431_020 |
0.1706386049 |
- |
- |
PREDICTED: F-box protein SKP2A [Jatropha curcas] |
| 11 |
Hb_106321_030 |
0.1712426094 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 12 |
Hb_008725_150 |
0.1760306938 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 13 |
Hb_048476_170 |
0.1764236649 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |
| 14 |
Hb_165350_010 |
0.1778608811 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 15 |
Hb_001247_200 |
0.1789707826 |
- |
- |
PREDICTED: clathrin coat assembly protein AP180 [Jatropha curcas] |
| 16 |
Hb_004221_050 |
0.1834024114 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g01540 [Jatropha curcas] |
| 17 |
Hb_000755_080 |
0.1851962058 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 18 |
Hb_000270_780 |
0.1866740708 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 19 |
Hb_011457_160 |
0.1868814191 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 20 |
Hb_002701_080 |
0.1880873121 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like [Jatropha curcas] |