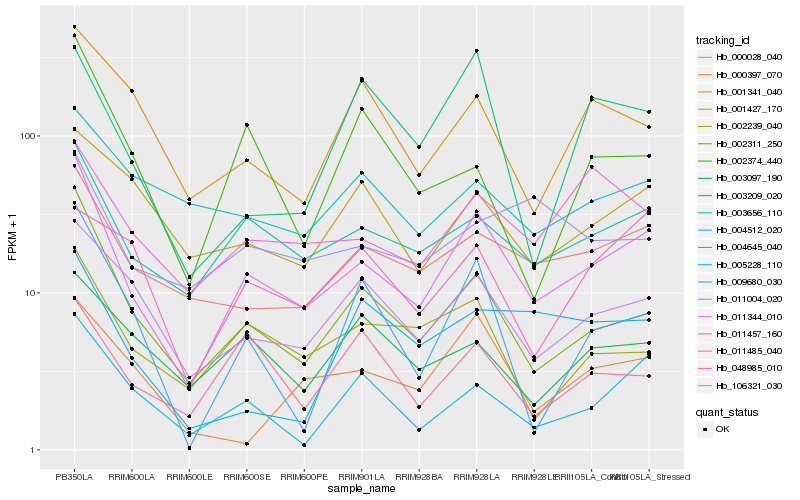
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011457_160 |
0.0 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 2 |
Hb_002311_250 |
0.1369113999 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 3 |
Hb_003656_110 |
0.1412394319 |
- |
- |
phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000028_040 |
0.1595746579 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |
| 5 |
Hb_011344_010 |
0.1635458699 |
- |
- |
PREDICTED: NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 [Jatropha curcas] |
| 6 |
Hb_004645_040 |
0.1856568878 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 7 |
Hb_002239_040 |
0.1868814191 |
- |
- |
Cellular apoptosis susceptibility protein / importin-alpha re-exporter, putative isoform 1 [Theobroma cacao] |
| 8 |
Hb_011485_040 |
0.1948653377 |
- |
- |
PREDICTED: probable mitochondrial adenine nucleotide transporter BTL3 [Jatropha curcas] |
| 9 |
Hb_001427_170 |
0.2004577511 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 10 |
Hb_000397_070 |
0.2034276635 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 11 |
Hb_001341_040 |
0.2041539672 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 12 |
Hb_009680_030 |
0.2061380616 |
- |
- |
PREDICTED: heat shock protein 83-like [Jatropha curcas] |
| 13 |
Hb_003097_190 |
0.2087270028 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 14 |
Hb_004512_020 |
0.2095691362 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 15 |
Hb_106321_030 |
0.2108200133 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 16 |
Hb_011004_020 |
0.2159755269 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 17 |
Hb_002374_440 |
0.2186667319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003209_020 |
0.2196725522 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 19 |
Hb_048985_010 |
0.2197199904 |
- |
- |
- |
| 20 |
Hb_005228_110 |
0.2232310252 |
- |
- |
Cleavage and polyadenylation specificity factor subunit 3 [Fukomys damarensis] |