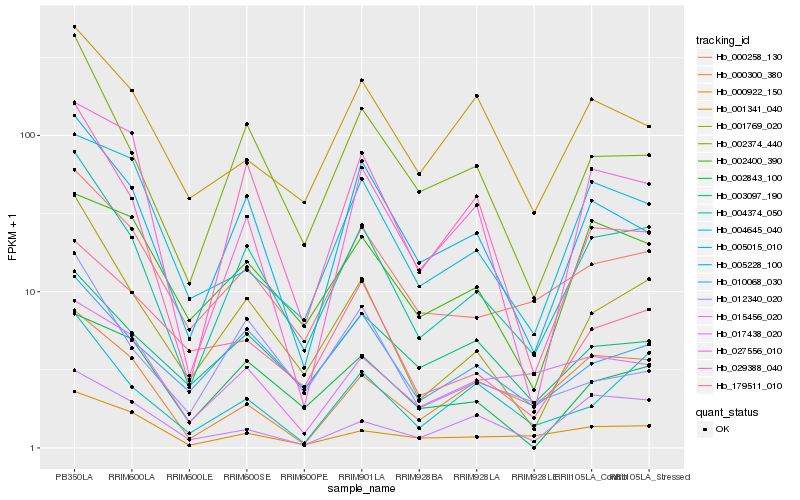
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001769_020 |
0.1010611266 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_002374_440 |
0.123577065 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005228_100 |
0.1307383091 |
- |
- |
- |
| 5 |
Hb_000258_130 |
0.1335413666 |
- |
- |
- |
| 6 |
Hb_010068_030 |
0.1505109177 |
- |
- |
aconitase, putative [Ricinus communis] |
| 7 |
Hb_004645_040 |
0.1541147399 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 8 |
Hb_179511_010 |
0.1573546092 |
- |
- |
- |
| 9 |
Hb_000300_380 |
0.1595874921 |
- |
- |
- |
| 10 |
Hb_000922_150 |
0.1604043096 |
- |
- |
- |
| 11 |
Hb_003097_190 |
0.1680130526 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 12 |
Hb_027556_010 |
0.1701909164 |
- |
- |
- |
| 13 |
Hb_002843_100 |
0.1721312437 |
- |
- |
- |
| 14 |
Hb_002400_390 |
0.1751295565 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 15 |
Hb_001341_040 |
0.1751385656 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 16 |
Hb_005015_010 |
0.1752558665 |
- |
- |
PREDICTED: programmed cell death protein 2-like [Jatropha curcas] |
| 17 |
Hb_017438_020 |
0.1757376543 |
- |
- |
- |
| 18 |
Hb_015456_020 |
0.1765776891 |
- |
- |
- |
| 19 |
Hb_012340_020 |
0.1779751781 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 20 |
Hb_029388_040 |
0.1797803483 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |