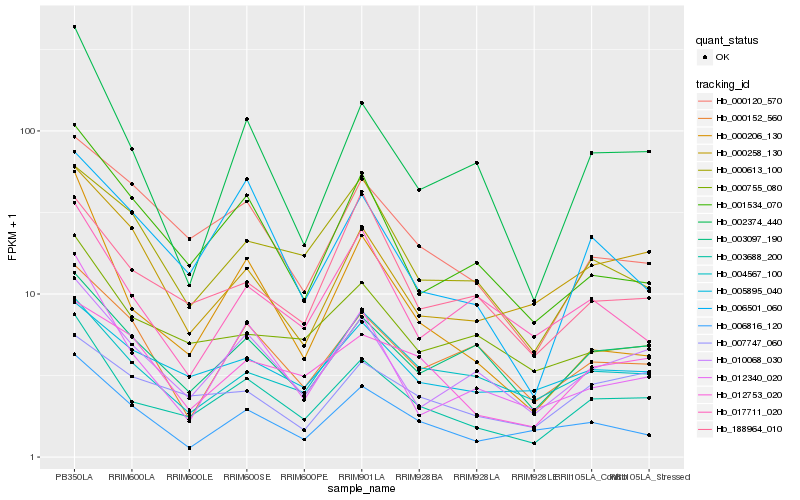
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_200 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010068_030 |
0.127382731 |
- |
- |
aconitase, putative [Ricinus communis] |
| 3 |
Hb_002374_440 |
0.1347640771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000120_570 |
0.1412503493 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 5 |
Hb_003097_190 |
0.1433109295 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 6 |
Hb_012340_020 |
0.1437324629 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000755_080 |
0.1457014665 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 8 |
Hb_001534_070 |
0.1459492603 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 9 |
Hb_000206_130 |
0.1494497177 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_006816_120 |
0.1514440981 |
- |
- |
Nicotianamine synthase, putative [Ricinus communis] |
| 11 |
Hb_006501_060 |
0.1555643229 |
- |
- |
PREDICTED: lachrymatory-factor synthase-like [Jatropha curcas] |
| 12 |
Hb_005895_040 |
0.1561798373 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 13 |
Hb_004567_100 |
0.1566378532 |
- |
- |
PREDICTED: uncharacterized protein LOC105644934 [Jatropha curcas] |
| 14 |
Hb_000258_130 |
0.1611163809 |
- |
- |
- |
| 15 |
Hb_012753_020 |
0.1623438394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000613_100 |
0.1629963369 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 8 [Jatropha curcas] |
| 17 |
Hb_000152_560 |
0.1638232763 |
- |
- |
hypothetical protein JCGZ_26236 [Jatropha curcas] |
| 18 |
Hb_007747_060 |
0.1654629581 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_017711_020 |
0.1655059047 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 56 isoform X1 [Jatropha curcas] |
| 20 |
Hb_188964_010 |
0.1655135743 |
- |
- |
maintenance of killer 16 (mak16) protein, putative [Ricinus communis] |