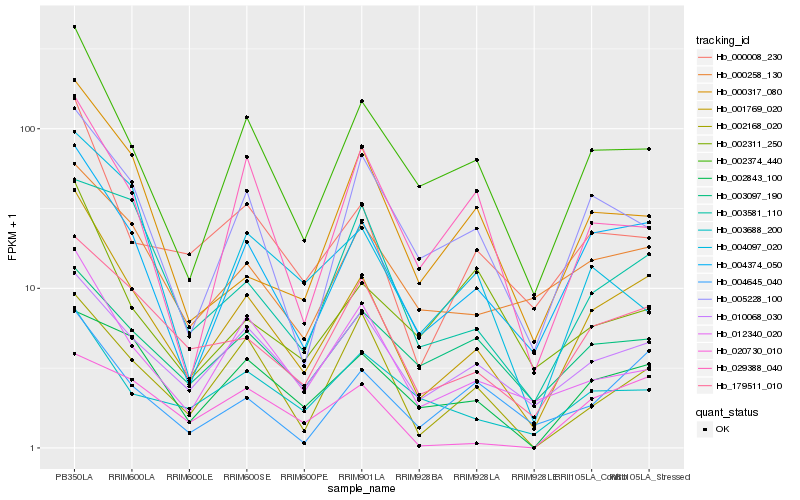
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001769_020 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_004374_050 |
0.1010611266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002374_440 |
0.1341910258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_179511_010 |
0.1463070796 |
- |
- |
- |
| 5 |
Hb_000008_230 |
0.1529366718 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 6 |
Hb_010068_030 |
0.1667419664 |
- |
- |
aconitase, putative [Ricinus communis] |
| 7 |
Hb_012340_020 |
0.168093467 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000258_130 |
0.1699018551 |
- |
- |
- |
| 9 |
Hb_005228_100 |
0.1734247186 |
- |
- |
- |
| 10 |
Hb_004645_040 |
0.1735763928 |
- |
- |
glucose-methanol-choline (gmc) oxidoreductase, putative [Ricinus communis] |
| 11 |
Hb_002843_100 |
0.1777564372 |
- |
- |
- |
| 12 |
Hb_003688_200 |
0.1787343205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000317_080 |
0.1841728129 |
- |
- |
PREDICTED: probable galacturonosyltransferase 10 [Jatropha curcas] |
| 14 |
Hb_002168_020 |
0.1948143688 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_029388_040 |
0.1970494199 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 16 |
Hb_003581_110 |
0.1981716436 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003097_190 |
0.2016316224 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 18 |
Hb_004097_020 |
0.2027766355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_020730_010 |
0.2066747524 |
- |
- |
hypothetical protein CICLE_v10010243mg, partial [Citrus clementina] |
| 20 |
Hb_002311_250 |
0.2077183485 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |