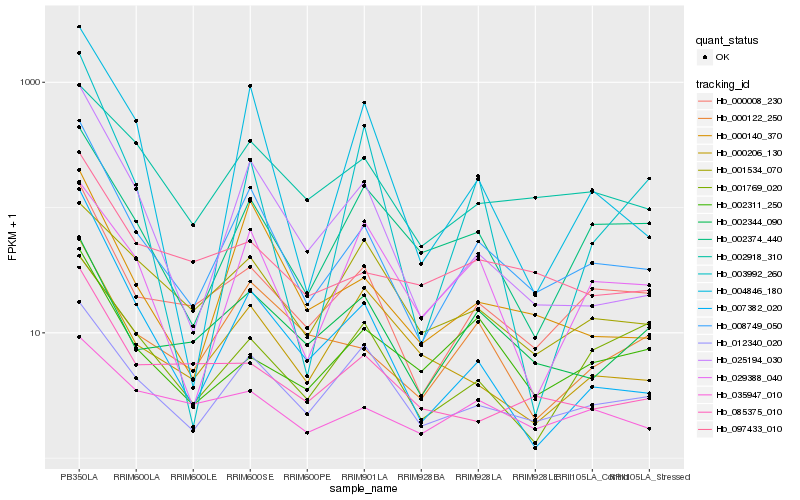
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008749_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 2 |
Hb_000008_230 |
0.1400953565 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 3 |
Hb_025194_030 |
0.1464903782 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 4 |
Hb_000140_370 |
0.1475537814 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6 [Jatropha curcas] |
| 5 |
Hb_012340_020 |
0.16043164 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 6 |
Hb_004846_180 |
0.1717982165 |
- |
- |
heat shock protein, putative [Ricinus communis] |
| 7 |
Hb_002918_310 |
0.1753067077 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 8 |
Hb_007382_020 |
0.1828479752 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 6-like [Jatropha curcas] |
| 9 |
Hb_002374_440 |
0.1847261995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_097433_010 |
0.1857292703 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 11 |
Hb_002344_090 |
0.1938966906 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 12 |
Hb_000206_130 |
0.1984680054 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_085375_010 |
0.2016071712 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_035947_010 |
0.2036935166 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 15 |
Hb_002311_250 |
0.2057432904 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 16 |
Hb_029388_040 |
0.2089067206 |
- |
- |
PREDICTED: GEM-like protein 5 [Jatropha curcas] |
| 17 |
Hb_001769_020 |
0.2100302213 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001534_070 |
0.2127328791 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 19 |
Hb_000122_250 |
0.2163381063 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 2C, putative [Ricinus communis] |
| 20 |
Hb_003992_260 |
0.2178497701 |
- |
- |
PREDICTED: 26.5 kDa heat shock protein, mitochondrial [Jatropha curcas] |