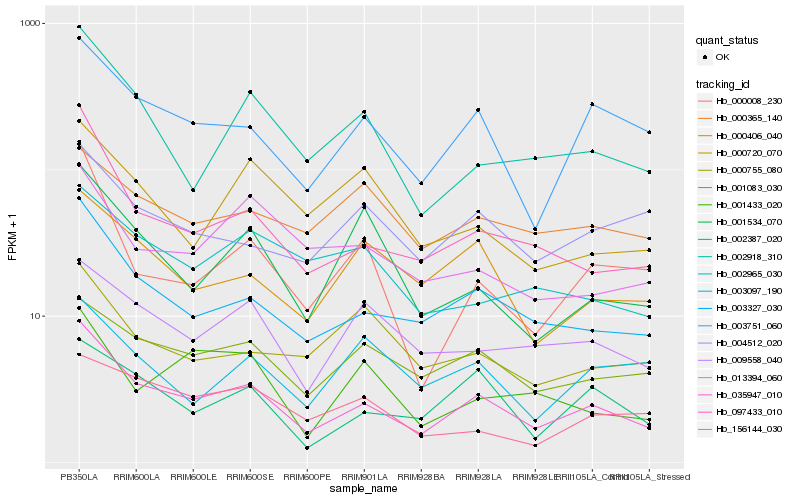
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035947_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 2 |
Hb_003327_030 |
0.1034460404 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 3 |
Hb_097433_010 |
0.1244658522 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 4 |
Hb_003751_060 |
0.1297384431 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 5 |
Hb_002918_310 |
0.143447168 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 6 |
Hb_001083_030 |
0.1511127052 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 7 |
Hb_002387_020 |
0.1538892603 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 8 |
Hb_009558_040 |
0.1571214113 |
transcription factor |
TF Family: MYB |
hypothetical protein PHAVU_008G102000g [Phaseolus vulgaris] |
| 9 |
Hb_000406_040 |
0.1610410986 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: FRIGIDA-like protein 5 [Jatropha curcas] |
| 10 |
Hb_000008_230 |
0.165088908 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 11 |
Hb_001534_070 |
0.1703463533 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 12 |
Hb_002965_030 |
0.1727220989 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 13 |
Hb_013394_060 |
0.1731509321 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 14 |
Hb_004512_020 |
0.1732944448 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 15 |
Hb_000755_080 |
0.1768730418 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 16 |
Hb_001433_020 |
0.1783506331 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 17 |
Hb_156144_030 |
0.1789121771 |
- |
- |
hypothetical protein RCOM_0187400 [Ricinus communis] |
| 18 |
Hb_000365_140 |
0.1808209735 |
- |
- |
PREDICTED: ethanolamine-phosphate cytidylyltransferase [Jatropha curcas] |
| 19 |
Hb_000720_070 |
0.1821758245 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_20639 [Jatropha curcas] |
| 20 |
Hb_003097_190 |
0.1828666843 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |