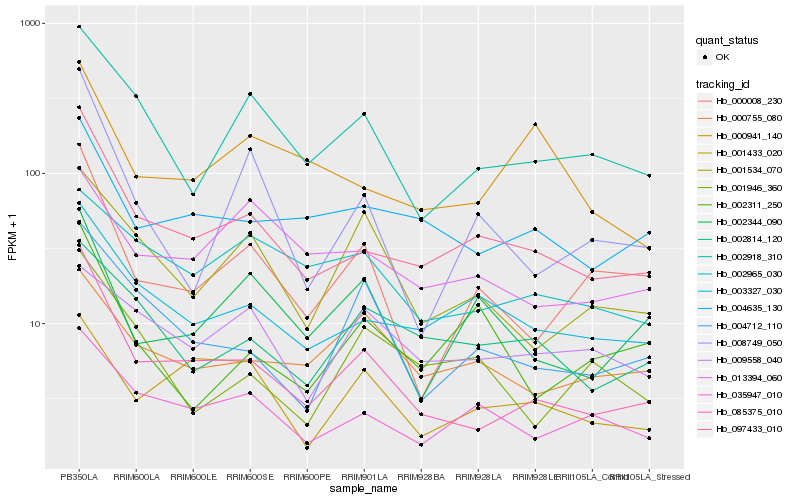
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097433_010 |
0.0 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TDX [Jatropha curcas] |
| 2 |
Hb_003327_030 |
0.0844108453 |
- |
- |
Hsc70-interacting protein, putative [Ricinus communis] |
| 3 |
Hb_035947_010 |
0.1244658522 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 4 |
Hb_085375_010 |
0.1455473137 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_002918_310 |
0.1634155144 |
- |
- |
PREDICTED: dnaJ protein homolog 1-like [Jatropha curcas] |
| 6 |
Hb_004635_130 |
0.1658561672 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 7 |
Hb_002814_120 |
0.1711149304 |
- |
- |
PREDICTED: uncharacterized protein LOC105633463 [Jatropha curcas] |
| 8 |
Hb_000008_230 |
0.1746501925 |
- |
- |
hypothetical protein POPTR_0004s22120g [Populus trichocarpa] |
| 9 |
Hb_000941_140 |
0.1851747814 |
- |
- |
PREDICTED: activator of 90 kDa heat shock protein ATPase homolog 1 [Jatropha curcas] |
| 10 |
Hb_008749_050 |
0.1857292703 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C4H3.01 [Jatropha curcas] |
| 11 |
Hb_002311_250 |
0.1870907273 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 12 |
Hb_013394_060 |
0.1939898479 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 13 |
Hb_002344_090 |
0.1946499653 |
- |
- |
PREDICTED: uncharacterized protein LOC105637976 [Jatropha curcas] |
| 14 |
Hb_004712_110 |
0.1990771048 |
- |
- |
30S ribosomal protein S10, putative [Ricinus communis] |
| 15 |
Hb_001946_360 |
0.1993017176 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 16 |
Hb_000755_080 |
0.2004174247 |
- |
- |
hypothetical protein RCOM_1016710 [Ricinus communis] |
| 17 |
Hb_002965_030 |
0.2048232814 |
- |
- |
PREDICTED: uncharacterized protein LOC105643493 [Jatropha curcas] |
| 18 |
Hb_001433_020 |
0.2102749097 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 19 |
Hb_001534_070 |
0.2126435313 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Eucalyptus grandis] |
| 20 |
Hb_009558_040 |
0.2139041512 |
transcription factor |
TF Family: MYB |
hypothetical protein PHAVU_008G102000g [Phaseolus vulgaris] |